4 An Introduction To Prokaryotic Cells And Microorganisms
4.1 Basic Characteristics Of Life
In the past, there have been many attempts to define what is meant by “life” through obsolete concepts such as odic force, hylomorphism, spontaneous generation and vitalism, that have now been disproved by biological discoveries. Aristotle is considered to be the first person to classify organisms. Later, Carl Linnaeus introduced his system of binomial nomenclature for the classification of species. Eventually new groups and categories of life were discovered, such as cells and microorganisms, forcing dramatic revisions of the structure of relationships between living organisms. Though currently only known on Earth, life need not be restricted to it, and many scientists speculate in the existence of extraterrestrial life. Artificial life is a computer simulation or human-made reconstruction of any aspect of life, which is often used to examine systems related to natural life.
Death is the permanent termination of all biological functions which sustain an organism, and as such, is the end of its life. Extinction is the term describing the dying out of a group or taxon, usually a species. Fossils are the preserved remains or traces of organisms.
The definition of life has long been a challenge for scientists and philosophers, with many varied definitions put forward. This is partially because life is a process, not a substance. This is complicated by a lack of knowledge of the characteristics of living entities, if any, that may have developed outside of Earth. Philosophical definitions of life have also been put forward, with similar difficulties on how to distinguish living things from the non-living. Legal definitions of life have also been described and debated, though these generally focus on the decision to declare a human dead, and the legal ramifications of this decision.
Since there is no unequivocal definition of life, most current definitions in biology are descriptive. Life is considered a characteristic of something that preserves, furthers or reinforces its existence in the given environment. This characteristic exhibits all or most of the following traits:
- Homeostasis: regulation of the internal environment to maintain a constant state; for example, sweating to reduce temperature
- Organization: being structurally composed of one or more cells – the basic units of life
- Metabolism: transformation of energy by converting chemicals and energy into cellular components (anabolism) and decomposing organic matter (catabolism). Living things require energy to maintain internal organization (homeostasis) and to produce the other phenomena associated with life.
- Growth: maintenance of a higher rate of anabolism than catabolism. A growing organism increases in size in all of its parts, rather than simply accumulating matter.
- Adaptation: the ability to change over time in response to the environment. This ability is fundamental to the process of evolution and is determined by the organism’s heredity, diet, and external factors.
- Response to stimuli: a response can take many forms, from the contraction of a unicellular organism to external chemicals, to complex reactions involving all the senses of multicellular organisms. A response is often expressed by motion; for example, the leaves of a plant turning toward the sun (phototropism), and chemotaxis.
- Reproduction: the ability to produce new individual organisms, either asexually from a single parent organism or sexually from two parent organisms.
These complex processes, called physiological functions, have underlying physical and chemical bases, as well as signaling and control mechanisms that are essential to maintaining life.
More than 99% of all species of life forms, amounting to over five billion species, that ever lived on Earth are estimated to be extinct.
Although the number of Earth’s catalogued species of lifeforms is between 1.2 million and 2 million, the total number of species in the planet is uncertain. Estimates range from 8 million to 100 million, with a more narrow range between 10 and 14 million, but it may be as high as 1 trillion (with only one-thousandth of one percent of the species described) according to studies realized in May 2016. The total number of related DNA base pairs on Earth is estimated at 5.0 x 1037 and weighs 50 billion tonnes. In comparison, the total mass of the biosphere has been estimated to be as much as 4 TtC (trillion tons of carbon). In July 2016, scientists reported identifying a set of 355 genes from the Last Universal Common Ancestor (LUCA) of all organisms living on Earth.
4.2 Origin Of Life
The Ancient Greeks believed that living things could spontaneously come into being from nonliving matter, and that the goddess Gaia could make life arise spontaneously from stones – a process known as Generatio spontanea. Aristotle disagreed, but he still believed that creatures could arise from dissimilar organisms or from soil. Variations of this concept of spontaneous generation still existed as late as the 17th century, but towards the end of the 17th century, a series of observations and arguments began that eventually discredited such ideas. This advance in scientific understanding was met with much opposition, with personal beliefs and individual prejudices often obscuring the facts.
William Harvey (1578–1657) was an early proponent of all life beginning from an egg, omne vivum ex ovo. Francesco Redi, an Italian physician, proved as early as 1668 that higher forms of life did not originate spontaneously by demonstrating that maggots come from eggs of flies. But proponents of spontaneous generation claimed that this did not apply to microbes and continued to hold that these could arise spontaneously. Attempts to disprove the spontaneous generation of life from non-life continued in the early 19th century with observations and experiments by Franz Schulze and Theodor Schwann. In 1745, John Needham added chicken broth to a flask and boiled it. He then let it cool and waited. Microbes grew, and he proposed it as an example of spontaneous generation. In 1768, Lazzaro Spallanzani repeated Needham’s experiment but removed all the air from the flask. No growth occurred. In 1854, Heinrich G. F. Schröder (1810–1885) and Theodor von Dusch, and in 1859, Schröder alone, repeated the Helmholtz filtration experiment and showed that living particles can be removed from air by filtering it through cotton-wool.
In 1864, Louis Pasteur finally announced the results of his scientific experiments. In a series of experiments similar to those performed earlier by Needham and Spallanzani, Pasteur demonstrated that life does not arise in areas that have not been contaminated by existing life. Pasteur’s empirical results were summarized in the phrase Omne vivum ex vivo, Latin for “all life [is] from life”.
All known life forms share fundamental molecular mechanisms, reflecting their common descent; based on these observations, hypotheses on the origin of life attempt to find a mechanism explaining the formation of a universal common ancestor, from simple organic molecules via pre-cellular life to protocells and metabolism. Models have been divided into “genes-first” and “metabolism-first” categories, but a recent trend is the emergence of hybrid models that combine both categories.
Life on Earth is based on carbon and water. Carbon provides stable frameworks for complex chemicals and can be easily extracted from the environment, especially from carbon dioxide. There is no other chemical element whose properties are similar enough to carbon’s to be called an analogue; silicon, the element directly below carbon on the periodic table, does not form very many complex stable molecules, and because most of its compounds are water-insoluble and because silicon dioxide is a hard and abrasive solid in contrast to carbon dioxide at temperatures associated with living things, it would be more difficult for organisms to extract. The elements boron and phosphorus have more complex chemistries, but suffer from other limitations relative to carbon. Water is an excellent solvent and has two other useful properties: the fact that ice floats enables aquatic organisms to survive beneath it in winter; and its molecules have electrically negative and positive ends, which enables it to form a wider range of compounds than other solvents can. Other good solvents, such as ammonia, are liquid only at such low temperatures that chemical reactions may be too slow to sustain life, and lack water’s other advantages. Organisms based on alternative biochemistry may, however, be possible on other planets.
Abiogenesis, or informally the origin of life, is the natural process of life arising from non-living matter, such as simple organic compounds. The prevailing scientific hypothesis is that the transition from non-living to living entities was not a single event, but a gradual process of increasing complexity. Although the occurrence of abiogenesis is uncontroversial among scientists, its possible mechanisms are poorly understood. There are several principles and hypotheses for how abiogenesis could have occurred. Life on Earth first appeared as early as 4.28 billion years ago, soon after ocean formation 4.41 billion years ago, and not long after the formation of the Earth 4.54 billion years ago. The earliest known life forms are microfossils of bacteria.
There is no current scientific consensus as to how life originated. However, many accepted scientific models build on the Miller–Urey experiment and the work of Sidney Fox, which show that conditions on the primitive Earth favored chemical reactions that synthesize amino acids and other organic compounds from inorganic precursors, and phospholipids spontaneously form lipid bilayers, the basic structure of a cell membrane.
The classic 1952 Miller–Urey experiment (Figure 4.1) demonstrated that most amino acids, the chemical constituents of the proteins used in all living organisms, can be synthesized from inorganic compounds under conditions intended to replicate those of the early Earth. The experiment used water (H2O), methane (CH4), ammonia (NH3), and hydrogen (H2). The chemicals were all sealed inside a sterile 5-liter glass flask connected to a 500 ml flask half-full of water. The water in the smaller flask was heated to induce evaporation, and the water vapour was allowed to enter the larger flask. Continuous electrical sparks were fired between the electrodes to simulate lightning in the water vapour and gaseous mixture, and then the simulated atmosphere was cooled again so that the water condensed and trickled into a U-shaped trap at the bottom of the apparatus.
After a day, the solution collected at the trap had turned pink in colour, and after a week of continuous operation the solution was deep red and turbid. The boiling flask was then removed, and mercuric chloride was added to prevent microbial contamination. The reaction was stopped by adding barium hydroxide and sulfuric acid, and evaporated to remove impurities. Using paper chromatography, Miller identified five amino acids present in the solution: glycine, α-alanine and β-alanine were positively identified, while aspartic acid and α-aminobutyric acid (AABA) were less certain, due to the spots being faint.Complex organic molecules occur in the Solar System and in interstellar space, and these molecules may have provided starting material for the development of life on Earth.
Figure 4.1: The Miller–Urey experiment was a chemical experiment that simulated the conditions thought at the time (1952) to be present on the early Earth and tested the chemical origin of life under those conditions. The experiment at the time supported Alexander Oparin’s and J. B. S. Haldane’s hypothesis that putative conditions on the primitive Earth favoured chemical reactions that synthesized more complex organic compounds from simpler inorganic precursors. Considered to be the classic experiment investigating abiogenesis, it was conducted in 1952 by Stanley Miller, with assistance from Harold Urey, at the University of Chicago and later the University of California, San Diego and published the following year.
Living organisms synthesize proteins, which are polymers of amino acids using instructions encoded by deoxyribonucleic acid (DNA). Protein synthesis entails intermediary ribonucleic acid (RNA) polymers. One possibility for how life began is that genes originated first, followed by proteins; the alternative being that proteins came first and then genes.
However, because genes and proteins are both required to produce the other, the problem of considering which came first is like that of the chicken or the egg. Most scientists have adopted the hypothesis that because of this, it is unlikely that genes and proteins arose independently.
Therefore, a possibility, first suggested by Francis Crick, is that the first life was based on RNA, which has the DNA-like properties of information storage and the catalytic properties of some proteins. This is called the RNA world hypothesis, and it is supported by the observation that many of the most critical components of cells (those that evolve the slowest) are composed mostly or entirely of RNA. Also, many critical cofactors (ATP, Acetyl-CoA, NADH, etc.) are either nucleotides or substances clearly related to them. The catalytic properties of RNA had not yet been demonstrated when the hypothesis was first proposed, but they were confirmed by Thomas Cech in 1986.
One issue with the RNA world hypothesis is that synthesis of RNA from simple inorganic precursors is more difficult than for other organic molecules. One reason for this is that RNA precursors are very stable and react with each other very slowly under ambient conditions, and it has also been proposed that living organisms consisted of other molecules before RNA. However, the successful synthesis of certain RNA molecules under the conditions that existed prior to life on Earth has been achieved by adding alternative precursors in a specified order with the precursor phosphate present throughout the reaction. This study makes the RNA world hypothesis more plausible.
Geological findings in 2013 showed that reactive phosphorus species (like phosphite) were in abundance in the ocean before 3.5 Ga, and that Schreibersite easily reacts with aqueous glycerol to generate phosphite and glycerol 3-phosphate. It is hypothesized that Schreibersite-containing meteorites from the Late Heavy Bombardment could have provided early reduced phosphorus, which could react with prebiotic organic molecules to form phosphorylated biomolecules, like RNA.
In 2009, experiments demonstrated Darwinian evolution of a two-component system of RNA enzymes (ribozymes) in vitro. The work was performed in the laboratory of Gerald Joyce, who stated “This is the first example, outside of biology, of evolutionary adaptation in a molecular genetic system.”
Prebiotic compounds may have originated extraterrestrially. NASA findings in 2011, based on studies with meteorites found on Earth, suggest DNA and RNA components (adenine, guanine and related organic molecules) may be formed in outer space.
In March 2015, NASA scientists reported that, for the first time, complex DNA and RNA organic compounds of life, including uracil, cytosine and thymine, have been formed in the laboratory under outer space conditions, using starting chemicals, such as pyrimidine, found in meteorites. Pyrimidine, like polycyclic aromatic hydrocarbons (PAHs), the most carbon-rich chemical found in the universe, may have been formed in red giants or in interstellar dust and gas clouds, according to the scientists.
According to the panspermia hypothesis, microscopic life—distributed by meteoroids, asteroids and other small Solar System bodies—may exist throughout the universe.
Since its primordial beginnings, life on Earth has changed its environment on a geologic time scale, but it has also adapted to survive in most ecosystems and conditions. Some microorganisms, called extremophiles, thrive in physically or geochemically extreme environments that are detrimental to most other life on Earth. The cell is considered the structural and functional unit of life. There are two kinds of cells, prokaryotic and eukaryotic, both of which consist of cytoplasm enclosed within a membrane and contain many biomolecules such as proteins and nucleic acids. Cells reproduce through a process of cell division, in which the parent cell divides into two or more daughter cells.
Figure 4.2: Cartoons of a eukaryotic and prokaryotic cell.
4.3 The Foundations Of Modern Biology
4.3.1 The Cell Theory
Cell theory states that the cell is the fundamental unit of life, that all living things are composed of one or more cells, and that all cells arise from pre-existing cells through cell division. In multicellular organisms, every cell in the organism’s body derives ultimately from a single cell in a fertilized egg. The cell is also considered to be the basic unit in many pathological processes. In addition, the phenomenon of energy flow occurs in cells in processes that are part of the function known as metabolism. Finally, cells contain hereditary information (DNA), which is passed from cell to cell during cell division. Research into the origin of life, abiogenesis, amounts to an attempt to discover the origin of the first cells.
Cells are the basic unit of structure in every living thing, and all cells arise from pre-existing cells by division. Cell theory was formulated by Henri Dutrochet, Theodor Schwann, Matthias Jakob Schleiden, Rudolf Virchow and others during the early nineteenth century, and subsequently became widely accepted. The activity of an organism depends on the total activity of its cells, with energy flow occurring within and between them. Cells contain hereditary information that is carried forward as a genetic code during cell division.
There are two primary types of cells. Prokaryotes lack a nucleus and other membrane-bound organelles, although they have circular DNA and ribosomes. Bacteria and Archaea are two domains of prokaryotes. The other primary type of cells are the eukaryotes, which have distinct nuclei bound by a nuclear membrane and membrane-bound organelles, including mitochondria, chloroplasts, lysosomes, rough and smooth endoplasmic reticulum, and vacuoles. In addition, they possess organized chromosomes that store genetic material. All species of large complex organisms are eukaryotes, including animals, plants and fungi, though most species of eukaryote are protist microorganisms. The conventional model is that eukaryotes evolved from prokaryotes, with the main organelles of the eukaryotes forming through endosymbiosis between bacteria and the progenitor eukaryotic cell.
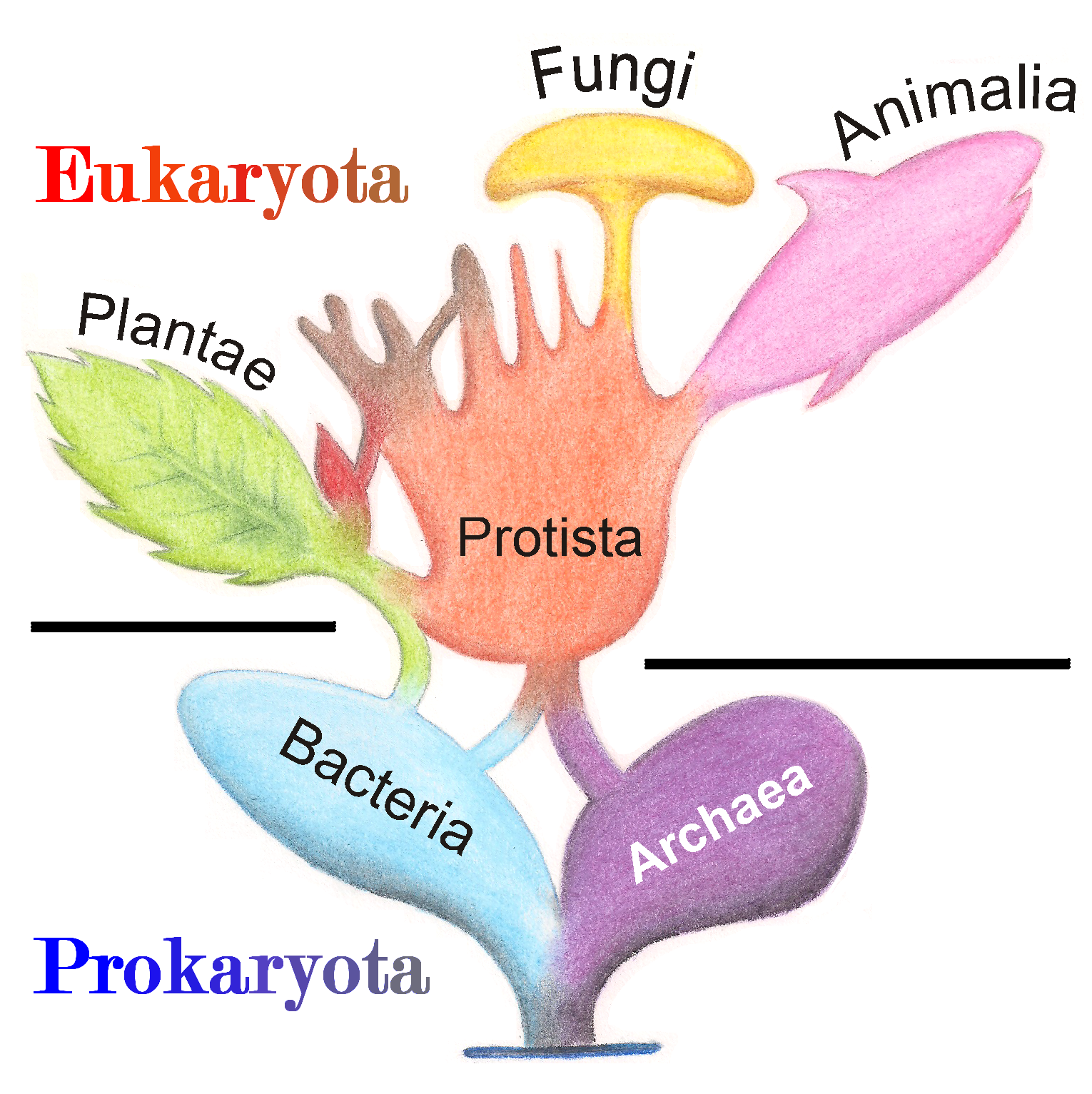
Figure 4.3: Tree diagram illustrating the evolutionary relationship of living organisms.
A virus is a submicroscopic infectious agent that replicates only inside the living cells of an organism. Scientific opinions differ on whether viruses are a form of life, or organic structures that interact with living organisms. They have been described as “organisms at the edge of life”, since they resemble organisms in that they possess genes, evolve by natural selection, and reproduce by creating multiple copies of themselves through self-assembly. Although they have genes, they do not have a cellular structure, which is seen as the basic unit of life. Viruses do not have their own metabolism, and require a host cell to make new products. They therefore cannot naturally reproduce outside a host cell—although bacterial species such as rickettsia and chlamydia are considered living organisms despite the same limitation. Accepted forms of life use cell division to reproduce, whereas viruses spontaneously assemble within cells. They differ from autonomous growth of crystals as they inherit genetic mutations while being subject to natural selection. Virus self-assembly within host cells has implications for the study of the origin of life, as it lends further credence to the hypothesis that life could have started as self-assembling organic molecules.
The molecular mechanisms of cell biology are based on proteins which are synthesized by the ribosomes through an enzyme-catalyzed process called protein biosynthesis. A sequence of amino acids is assembled and joined together based upon gene expression of the cell’s nucleic acid. In eukaryotic cells, these proteins may then be transported and processed through the Golgi apparatus in preparation for dispatch to their destination.
Cells reproduce through a process of cell division in which the parent cell divides into two or more daughter cells. For prokaryotes, cell division occurs through a process of fission in which the DNA is replicated, then the two copies are attached to parts of the cell membrane. In eukaryotes, a more complex process of mitosis is followed. However, the end result is the same; the resulting cell copies are identical to each other and to the original cell (except for mutations), and both are capable of further division following an interphase period.
Multicellular organisms may have first evolved through the formation of colonies of identical cells. These cells can form group organisms through cell adhesion. The individual members of a colony are capable of surviving on their own, whereas the members of a true multi-cellular organism have developed specializations, making them dependent on the remainder of the organism for survival. Such organisms are formed clonally or from a single germ cell that is capable of forming the various specialized cells that form the adult organism. This specialization allows multicellular organisms to exploit resources more efficiently than single cells. In January 2016, scientists reported that, about 800 million years ago, a minor genetic change in a single molecule, called GK-PID, may have allowed organisms to go from a single cell organism to one of many cells.
Cells have evolved methods to perceive and respond to their microenvironment, thereby enhancing their adaptability. Cell signaling coordinates cellular activities, and hence governs the basic functions of multicellular organisms. Signaling between cells can occur through direct cell contact using juxtacrine signalling, or indirectly through the exchange of agents as in the endocrine system. In more complex organisms, coordination of activities can occur through a dedicated nervous system.
4.3.2 The Theory Of Evolution
A central organizing concept in biology is that life changes and develops through evolution, and that all life-forms known have a common origin. The theory of evolution postulates that all organisms on the Earth, both living and extinct, have descended from a common ancestor or an ancestral gene pool. This universal common ancestor of all organisms is believed to have appeared about 3.5 billion years ago. Biologists regard the ubiquity of the genetic code as definitive evidence in favor of the theory of universal common descent for all bacteria, archaea, and eukaryotes.
The term “evolution” was introduced into the scientific lexicon by Jean-Baptiste de Lamarck in 1809, and fifty years later Charles Darwin posited a scientific model of natural selection as evolution’s driving force. Alfred Russel Wallace independently reached the same conclusions and is recognized as the co-discoverer of this concept. Evolution is now used to explain the great variations of life found on Earth.
Darwin theorized that species flourish or die when subjected to the processes of natural selection or selective breeding. Genetic drift was embraced as an additional mechanism of evolutionary development in the modern synthesis of the theory.
The evolutionary history of the species—which describes the characteristics of the various species from which it descended—together with its genealogical relationship to every other species is known as its phylogeny. Widely varied approaches to biology generate information about phylogeny. These include the comparisons of DNA sequences, a product of molecular biology (more particularly genomics), and comparisons of fossils or other records of ancient organisms, a product of paleontology. Biologists organize and analyze evolutionary relationships through various methods, including phylogenetics, phenetics, and cladistics
Evolution is relevant to the understanding of the natural history of life forms and to the understanding of the organization of current life forms. But, those organizations can only be understood in light of how they came to be by way of the process of evolution. Consequently, evolution is central to all fields of biology.
The evolutionary history of life on Earth traces the processes by which living and fossil organisms evolved, from the earliest emergence of life to the present. Earth formed about 4.5 billion years (Ga) ago and evidence suggests life emerged prior to 3.7 Ga. (Although there is some evidence of life as early as 4.1 to 4.28 Ga, it remains controversial due to the possible non-biological formation of the purported fossils.) The similarities among all known present-day species indicate that they have diverged through the process of evolution from a common ancestor. Approximately 1 trillion species currently live on Earth of which only 1.75–1.8 million have been named and 1.6 million documented in a central database. These currently living species represent less than one percent of all species that have ever lived on earth.
The earliest evidence of life comes from biogenic carbon signatures and stromatolite fossils discovered in 3.7 billion-year-old metasedimentary rocks from western Greenland. In 2015, possible “remains of biotic life” were found in 4.1 billion-year-old rocks in Western Australia. In March 2017, putative evidence of possibly the oldest forms of life on Earth was reported in the form of fossilized microorganisms discovered in hydrothermal vent precipitates in the Nuvvuagittuq Belt of Quebec, Canada, that may have lived as early as 4.28 billion years ago, not long after the oceans formed 4.4 billion years ago, and not long after the formation of the Earth 4.54 billion years ago.
Microbial mats of coexisting bacteria and archaea were the dominant form of life in the early Archean Epoch and many of the major steps in early evolution are thought to have taken place in this environment. The evolution of photosynthesis, around 3.5 Ga, eventually led to a buildup of its waste product, oxygen, in the atmosphere, leading to the great oxygenation event, beginning around 2.4 Ga. The earliest evidence of eukaryotes (complex cells with organelles) dates from 1.85 Ga, and while they may have been present earlier, their diversification accelerated when they started using oxygen in their metabolism. Later, around 1.7 Ga, multicellular organisms began to appear, with differentiated cells performing specialised functions. Sexual reproduction, which involves the fusion of male and female reproductive cells (gametes) to create a zygote in a process called fertilization is, in contrast to asexual reproduction, the primary method of reproduction for the vast majority of macroscopic organisms, including almost all eukaryotes (which includes animals and plants). However the origin and evolution of sexual reproduction remain a puzzle for biologists though it did evolve from a common ancestor that was a single celled eukaryotic species. Bilateria, animals having a left and a right side that are mirror images of each other, appeared by 555 Ma (million years ago).
The earliest plants on land date back to around 850 million years ago (Ma), from carbon isotopes in Precambrian rocks, while algae-like multicellular land plants are dated back even to about 1 billion years ago, although evidence suggests that microorganisms formed the earliest terrestrial ecosystems, at least 2.7 billion years ago (Ga). Microorganisms are thought to have paved the way for the inception of land plants in the Ordovician. Land plants were so successful that they are thought to have contributed to the Late Devonian extinction event. (The long causal chain implied seems to involve the success of early tree archaeopteris (1) drew down CO2 levels, leading to global cooling and lowered sea levels, (2) roots of archeopteris fostered soil development which increased rock weathering, and the subsequent nutrient run-off may have triggered algal blooms resulting in anoxic events which caused marine-life die-offs. Marine species were the primary victims of the Late Devonian extinction.)
Ediacara biota appear during the Ediacaran period, while vertebrates, along with most other modern phyla originated about 525 Ma during the Cambrian explosion. During the Permian period, synapsids, including the ancestors of mammals, dominated the land, but most of this group became extinct in the Permian–Triassic extinction event 252 Ma. During the recovery from this catastrophe, archosaurs became the most abundant land vertebrates; one archosaur group, the dinosaurs, dominated the Jurassic and Cretaceous periods. After the Cretaceous–Paleogene extinction event 65 Ma killed off the non-avian dinosaurs, mammals increased rapidly in size and diversity. Such mass extinctions may have accelerated evolution by providing opportunities for new groups of organisms to diversify.
4.3.3 Genetics
Genes are the primary units of inheritance in all organisms. A gene is a unit of heredity and corresponds to a region of DNA that influences the form or function of an organism in specific ways. All organisms, from bacteria to animals, share the same basic machinery that copies and translates DNA into proteins. Cells transcribe a DNA gene into an RNA version of the gene, and a ribosome then translates the RNA into a sequence of amino acids known as a protein. The translation code from RNA codon to amino acid is the same for most organisms. For example, a sequence of DNA that codes for insulin in humans also codes for insulin when inserted into other organisms, such as plants.
DNA is found as linear chromosomes in eukaryotes, and circular chromosomes in prokaryotes. A chromosome is an organized structure consisting of DNA and histones. The set of chromosomes in a cell and any other hereditary information found in the mitochondria, chloroplasts, or other locations is collectively known as a cell’s genome. In eukaryotes, genomic DNA is localized in the cell nucleus, or with small amounts in mitochondria and chloroplasts. In prokaryotes, the DNA is held within an irregularly shaped body in the cytoplasm called the nucleoid. The genetic information in a genome is held within genes, and the complete assemblage of this information in an organism is called its genotype.
4.3.4 Homeostasis
Homeostasis is the ability of an open system to regulate its internal environment to maintain stable conditions by means of multiple dynamic equilibrium adjustments that are controlled by interrelated regulation mechanisms. All living organisms, whether unicellular or multicellular, exhibit homeostasis.
To maintain dynamic equilibrium and effectively carry out certain functions, a system must detect and respond to perturbations. After the detection of a perturbation, a biological system normally responds through negative feedback that stabilize conditions by reducing or increasing the activity of an organ or system. One example is the release of glucagon when sugar levels are too low.
4.3.5 Energy
The survival of a living organism depends on the continuous input of energy. Chemical reactions that are responsible for its structure and function are tuned to extract energy from substances that act as its food and transform them to help form new cells and sustain them. In this process, molecules of chemical substances that constitute food play two roles; first, they contain energy that can be transformed and reused in that organism’s biological, chemical reactions; second, food can be transformed into new molecular structures (biomolecules) that are of use to that organism.
The organisms responsible for the introduction of energy into an ecosystem are known as producers or autotrophs. Nearly all such organisms originally draw their energy from the sun. Plants and other phototrophs use solar energy via a process known as photosynthesis to convert raw materials into organic molecules, such as ATP, whose bonds can be broken to release energy. A few ecosystems, however, depend entirely on energy extracted by chemotrophs from methane, sulfides, or other non-luminal energy sources.
Some of the energy thus captured produces biomass and energy that is available for growth and development of other life forms. The majority of the rest of this biomass and energy are lost as waste molecules and heat. The most important processes for converting the energy trapped in chemical substances into energy useful to sustain life are metabolism and cellular respiration.
Bacteria (singular bacterium) and Archaea (singular archaeon) constitute two domains of single-celled organisms. These microorganisms lack cell nuclei and are therefore prokaryotes.
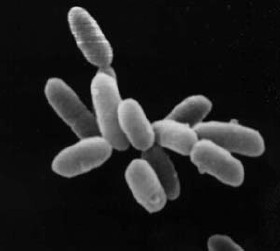
Scanning electron micrograph of Escherichia coli, grown in culture and adhered to a cover slip. Scanning electron micrograph of Escherichia coli bacteria.
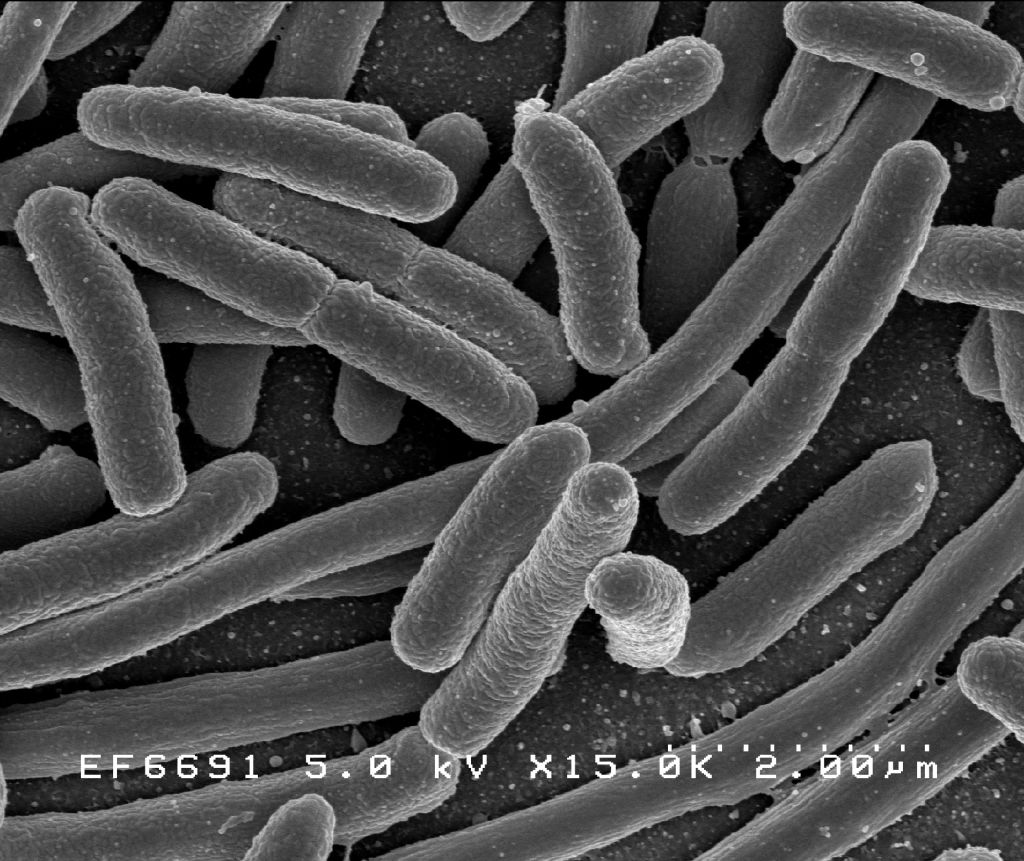
Figure 4.5: Scanning electron micrograph of Escherichia coli, grown in culture and adhered to a cover slip.
For much of the 20th century, prokaryotes were regarded as a single group of organisms and classified based on their biochemistry, morphology and metabolism. Microbiologists tried to classify microorganisms based on the structures of their cell walls, their shapes, and the substances they consume. In 1965, Emile Zuckerkandl and Linus Pauling instead proposed using the sequences of the genes in different prokaryotes to work out how they are related to each other. This phylogenetic approach is the main method used today.
Archaea – at that time only the methanogens were known – were first classified separately from bacteria in 1977 by Carl Woese and George E. Fox based on their ribosomal RNA (rRNA) genes. They called these groups the Urkingdoms of Archaebacteria and Eubacteria, though other researchers treated them as kingdoms or subkingdoms. Woese and Fox gave the first evidence for Archaebacteria as a separate “line of descent”: 1. lack of peptidoglycan in their cell walls, 2. two unusual coenzymes, 3. results of 16S ribosomal RNA gene sequencing. To emphasize this difference, Woese, Otto Kandler and Mark Wheelis later proposed reclassifying organisms into three natural domains known as the three-domain system: the Eukarya, the Bacteria and the Archaea, in what is now known as “The Woesian Revolution”.
4.4 Basic Characteristics Of Cells
The cell (from Latin cella, meaning “small room”) is the basic structural, functional, and biological unit of all known organisms. A cell is the smallest unit of life. Cells are often called the “building blocks of life”. The study of cells is called cell biology, cellular biology, or cytology.
Cells consist of cytoplasm enclosed within a membrane, which contains many biomolecules such as proteins and nucleic acids. Most plant and animal cells are only visible under a microscope, with dimensions between 1 and 100 micrometres. Organisms can be classified as unicellular (consisting of a single cell such as bacteria) or multicellular (including plants and animals). Most unicellular organisms are classed as microorganisms.
The number of cell in plants and animals varies from species to species; it has been estimated that humans contain somewhere around 40 trillion (4×1013) cells. The human brain accounts for around 80 billion of these cells.
In biology, cell theory is the historic scientific theory, now universally accepted, that living organisms are made up of cells, that they are the basic structural/organizational unit of all organisms, and that all cells come from pre-existing cells. Cells are the basic unit of structure in all organisms and also the basic unit of reproduction.
Credit for developing cell theory is usually given to two scientists: Theodor Schwann and Matthias Jakob Schleiden. While Rudolf Virchow contributed to the theory, he is not as credited for his attributions toward it. In 1839, Schleiden suggested that every structural part of a plant was made up of cells or the result of cells. He also suggested that cells were made by a crystallization process either within other cells or from the outside. However, this was not an original idea of Schleiden. He claimed this theory as his own, though Barthelemy Dumortier had stated it years before him. This crystallization process is no longer accepted with modern cell theory. In 1839, Theodor Schwann states that along with plants, animals are composed of cells or the product of cells in their structures. This was a major advancement in the field of biology since little was known about animal structure up to this point compared to plants. From these conclusions about plants and animals, two of the three tenets of cell theory were postulated.
- All living organisms are composed of one or more cells
- The cell is the most basic unit of life
Schleiden’s theory of free cell formation through crystallization was refuted in the 1850s by Robert Remak, Rudolf Virchow, and Albert von Kölliker. In 1855, Rudolf Virchow added the third tenet to cell theory. In Latin, this tenet states Omnis cellula e cellula. This translated to:
- All cells arise only from pre-existing cells
However, the idea that all cells come from pre-existing cells had in fact already been proposed by Robert Remak; it has been suggested that Virchow plagiarized Remak and did not give him credit. Remak published observations in 1852 on cell division, claiming Schleiden and Schawnn were incorrect about generation schemes. He instead said that binary fission, which was first introduced by Dumortier, was how reproduction of new animal cells were made. Once this tenet was added, the classical cell theory was complete.
The generally accepted parts of modern cell theory include:
- All living cells arise from pre-existing cells by division.
- The cell is the fundamental unit of structure and function in all living organisms.
- The activity of an organism depends on the total activity of independent cells.
- Energy flow (metabolism and biochemistry) occurs within cells.
- Cells contain DNA which is found specifically in the chromosome and RNA found in the cell nucleus and cytoplasm.
- All cells are basically the same in chemical composition in organisms of similar species.
The discovery of the cell was made possible through the invention of the microscope. In the first century BC, Romans were able to make glass, discovering that objects appeared to be larger under the glass. In Italy during the 12th century, Salvino D’Armate made a piece of glass fit over one eye, allowing for a magnification effect to that eye. The expanded use of lenses in eyeglasses in the 13th century probably led to wider spread use of simple microscopes (magnifying glasses) with limited magnification. Compound microscopes, which combine an objective lens with an eyepiece to view a real image achieving much higher magnification, first appeared in Europe around 1620. In 1665, Robert Hooke used a microscope about six inches long with two convex lenses inside and examined specimens under reflected light for the observations in his book Micrographia. Hooke also used a simpler microscope with a single lens for examining specimens with directly transmitted light, because this allowed for a clearer image.

Figure 4.7: Title page of “MICROGRAPHIA or some physiological descriptions of minute bodies made by magnifying glasses with observations and inquiries thereupon”.
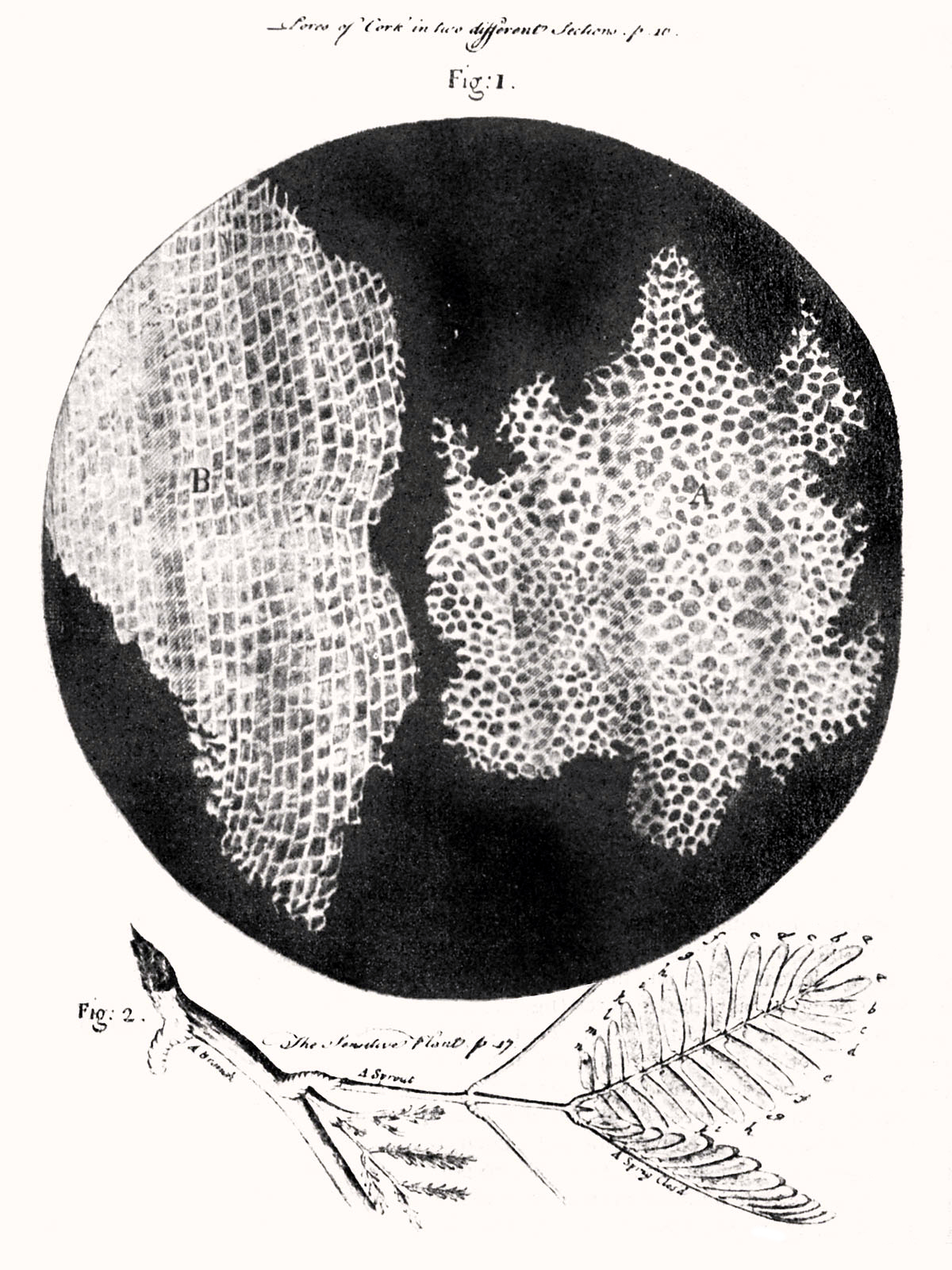
Figure 4.9: Hooke was the first to apply the word “cell”to biological objects. Cell structure of cork by Hooke.
Extensive microscopic study was done by Anton van Leeuwenhoek, a draper who took the interest in microscopes after seeing one while on an apprenticeship in Amsterdam in 1648. At some point in his life before 1668, he was able to learn how to grind lenses. This eventually led to Leeuwenhoek making his own unique microscope. His was a single lens simple microscope, rather than a compound microscope. This was because he was able to use a single lens that was a small glass sphere but allowed for a magnification of 270x. This was a large progression since the magnification before was only a maximum of 50x. After Leeuwenhoek, there was not much progress in microscope technology until the 1850s, two hundred years later. Carl Zeiss, a German engineer who manufactured microscopes, began to make changes to the lenses used. But the optical quality did not improve until the 1880s when he hired Otto Schott and eventually Ernst Abbe.
Optical microscopes can focus on objects the size of a wavelength or larger, giving restrictions still to advancement in discoveries with objects smaller than the wavelengths of visible light. The development of the electron microscope in the 1920s made it possible to view objects that are smaller than optical wavelengths, once again opening up new possibilities in science.
The cell was first discovered by Robert Hooke in 1665, which can be found to be described in his book Micrographia. In this book, he gave 60 ‘observations’ in detail of various objects under a coarse, compound microscope. One observation was from very thin slices of bottle cork. Hooke discovered a multitude of tiny pores that he named “cells”. This came from the Latin word Cella, meaning ‘a small room’ like monks lived in and also Cellulae, which meant the six sided cell of a honeycomb. However, Hooke did not know their real structure or function. What Hooke had thought were cells, were actually empty cell walls of plant tissues. With microscopes during this time having a low magnification, Hooke was unable to see that there were other internal components to the cells he was observing. Therefore, he did not think the “cellulae” were alive. His cell observations gave no indication of the nucleus and other organelles found in most living cells. In Micrographia, Hooke also observed mould, bluish in color, found on leather. After studying it under his microscope, he was unable to observe “seeds” that would have indicated how the mould was multiplying in quantity. This led to Hooke suggesting that spontaneous generation, from either natural or artificial heat, was the cause. Since this was an old Aristotelian theory still accepted at the time, others did not reject it and was not disproved until Leeuwenhoek later discovered that generation was achieved otherwise.
Anton van Leeuwenhoek is another scientist who saw these cells soon after Hooke did. He made use of a microscope containing improved lenses that could magnify objects almost 300-fold, or 270x. Under these microscopes, Leeuwenhoek found motile objects. In a letter to The Royal Society on October 9, 1676, he states that motility is a quality of life therefore these were living organisms. Over time, he wrote many more papers in which described many specific forms of microorganisms. Leeuwenhoek named these “animalcules,” which included protozoa and other unicellular organisms, like bacteria. Though he did not have much formal education, he was able to identify the first accurate description of red blood cells and discovered bacteria after gaining interest in the sense of taste that resulted in Leeuwenhoek to observe the tongue of an ox, then leading him to study “pepper water” in 1676. He also found for the first time the sperm cells of animals and humans. Once discovering these types of cells, Leeuwenhoek saw that the fertilization process requires the sperm cell to enter the egg cell. This put an end to the previous theory of spontaneous generation. After reading letters by Leeuwenhoek, Hooke was the first to confirm his observations that were thought to be unlikely by other contemporaries.
The cells in animal tissues were observed after plants were because the tissues were so fragile and susceptible to tearing, it was difficult for such thin slices to be prepared for studying. Biologists believed that there was a fundamental unit to life, but were unsure what this was. It would not be until over a hundred years later that this fundamental unit was connected to cellular structure and existence of cells in animals or plants. This conclusion was not made until Henri Dutrochet. Besides stating “the cell is the fundamental element of organization”, Dutrochet also claimed that cells were not just a structural unit, but also a physiological unit.
In 1804, Karl Rudolphi and Johann Heinrich Friedrich Link were awarded the prize for “solving the problem of the nature of cells”, meaning they were the first to prove that cells had independent cell walls by the Königliche Societät der Wissenschaft (Royal Society of Science), Göttingen. Before, it had been thought that cells shared walls and the fluid passed between them this way.
Cells can be subdivided into the following subcategories:
- Prokaryotes: Prokaryotes are relatively small cells surrounded by the plasma membrane, with a characteristic cell wall that may differ in composition depending on the particular organism. Prokaryotes lack a nucleus (although they do have circular or linear DNA) and other membrane-bound organelles (though they do contain ribosomes). The protoplasm of a prokaryote contains the chromosomal region that appears as fibrous deposits under the microscope, and the cytoplasm. Bacteria and Archaea are the two domains of prokaryotes.
- Eukaryotes: Eukaryotic cells are also surrounded by the plasma membrane, but on the other hand, they have distinct nuclei bound by a nuclear membrane or envelope. Eukaryotic cells also contain membrane-bound organelles, such as (mitochondria, chloroplasts, lysosomes, rough and smooth endoplasmic reticulum, vacuoles). In addition, they possess organized chromosomes which store genetic material. Animals have evolved a greater diversity of cell types in a multicellular body (100–150 different cell types), compared with 10–20 in plants, fungi, and protoctista.
The distinction between prokaryotes and eukaryotes was firmly established by the microbiologists Roger Stanier and C. B. van Niel in their 1962 paper The concept of a bacterium (though spelled procaryote and eucaryote there). That paper cites Édouard Chatton’s 1937 book Titres et Travaux Scientifiques for using those terms and recognizing the distinction. One reason for this classification was so that what was then often called blue-green algae (now called cyanobacteria) would not be classified as plants but grouped with bacteria.
4.5 Prokaryotic Cells
A prokaryote is a cellular organism that lacks an envelope-enclosed nucleus. The word prokaryote comes from the Greek πρό (pro, ‘before’) and κάρυον (karyon, ‘nut’ or ‘kernel’). In the two-empire system arising from the work of Édouard Chatton, prokaryotes were classified within the empire Prokaryota. But in the three-domain system, based upon molecular analysis, prokaryotes are divided into two domains: Bacteria (formerly Eubacteria) and Archaea (formerly Archaebacteria). Organisms with nuclei are placed in a third domain, Eukaryota. In the study of the origins of life, prokaryotes are thought to have arisen before eukaryotes.
Prokaryotes lack mitochondria, or any other eukaryotic membrane-bound organelles; and it was once thought that prokayotes lacked cellular compartments, and therefore all cellular components within the cytoplasm were unenclosed, except for an outer cell membrane. But bacterial microcompartments, which are thought to be primitive organelles enclosed in protein shells, have been discovered; and there is also evidence of prokayotic membrane-bound organelles. While typically being unicellular, some prokaryotes, such as cyanobacteria, may form large colonies. Others, such as myxobacteria, have multicellular stages in their life cycles. Prokaryotes are asexual, reproducing without fusion of gametes, although horizontal gene transfer also takes place.
Figure 4.10: Structure of a typical prokaryotic cell
Molecular studies have provided insight into the evolution and interrelationships of the three domains of life. The division between prokaryotes and eukaryotes reflects the existence of two very different levels of cellular organization; only eukaryotic cells have a enveloped nucleus that contains its chromosomal DNA, and other characteristic membrane-bound organelles including mitochondria. Distinctive types of prokaryotes include extremophiles and methanogens; these are common in some extreme environments.
Prokaryotes include bacteria and archaea, two of the three domains of life. Prokaryotic cells were the first form of life on Earth, characterized by having vital biological processes including cell signaling. They are simpler and smaller than eukaryotic cells, and lack a nucleus, and other membrane-bound organelles. The DNA of a prokaryotic cell consists of a single circular chromosome that is in direct contact with the cytoplasm. The nuclear region in the cytoplas is called the nucleoid. Most prokaryotes are the smallest of all organisms ranging from 0.5 to 2.0 µm in diameter.
A prokaryotic cell has three regions:
- Enclosing the cell is the cell envelope – generally consisting of a plasma membrane covered by a cell wall which, for some bacteria, may be further covered by a third layer called a capsule. Though most prokaryotes have both a cell membrane and a cell wall, there are exceptions such as Mycoplasma (bacteria) and Thermoplasma (archaea) which only possess the cell membrane layer. The envelope gives rigidity to the cell and separates the interior of the cell from its environment, serving as a protective filter. The cell wall consists of peptidoglycan in bacteria, and acts as an additional barrier against exterior forces. It also prevents the cell from expanding and bursting (cytolysis) from osmotic pressure due to a hypotonic environment. Some eukaryotic cells (plant cells and fungal cells) also have a cell wall.
- Inside the cell is the cytoplasmic region that contains the genome (DNA), ribosomes and various sorts of inclusions. The genetic material is freely found in the cytoplasm. Prokaryotes can carry extrachromosomal DNA elements called plasmids, which are usually circular. Linear bacterial plasmids have been identified in several species of spirochete bacteria, including members of the genus Borrelia notably Borrelia burgdorferi, which causes Lyme disease. Though not forming a nucleus, the DNA is condensed in a nucleoid. Plasmids encode additional genes, such as antibiotic resistance genes.
- On the outside, flagella and pili project from the cell’s surface. These are structures (not present in all prokaryotes) made of proteins that facilitate movement and communication between cells.
4.6 Bacteria
Bacteria (plural of the New Latin bacterium, which is the latinisation of the Greek βακτήριον (bakterion), the diminutive of βακτηρία (bakteria), meaning “staff, cane”, because the first ones to be discovered were rod-shaped) constitute a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria have a number of shapes, ranging from spheres to rods and spirals. Bacteria were among the first life forms to appear on Earth, and are present in most of its habitats. Bacteria inhabit soil, water, acidic hot springs, radioactive waste, and the deep biosphere of the earth’s crust. Bacteria also live in symbiotic and parasitic relationships with plants and animals. Most bacteria have not been characterised, and only about 27 percent of the bacterial phyla have species that can be grown in the laboratory. The study of bacteria is known as bacteriology, a branch of microbiology.
Nearly all animal life is dependent on bacteria for survival as only bacteria and some archaea possess the genes and enzymes necessary to synthesize vitamin B12, also known as cobalamin, and provide it through the food chain. Vitamin B12 is a water-soluble vitamin that is involved in the metabolism of every cell of the human body. It is a cofactor in DNA synthesis, and in both fatty acid and amino acid metabolism. It is particularly important in the normal functioning of the nervous system via its role in the synthesis of myelin.
There are typically 40 million bacterial cells in a gram of soil and a million bacterial cells in a millilitre of fresh water. There are approximately 5×1030 bacteria on Earth, forming a biomass which exceeds that of all plants and animals. Bacteria are vital in many stages of the nutrient cycle by recycling nutrients such as the fixation of nitrogen from the atmosphere. The nutrient cycle includes the decomposition of dead bodies; bacteria are responsible for the putrefaction stage in this process. In the biological communities surrounding hydrothermal vents and cold seeps, extremophile bacteria provide the nutrients needed to sustain life by converting dissolved compounds, such as hydrogen sulphide and methane, to energy.
In humans and most animals the largest number of bacteria exist in the gut, and a large number on the skin. The vast majority of the bacteria in the body are rendered harmless by the protective effects of the immune system, though many are beneficial, particularly in the gut flora. However, several species of bacteria are pathogenic and cause infectious diseases, including cholera, syphilis, anthrax, leprosy, and bubonic plague. The most common fatal bacterial diseases are respiratory infections. Tuberculosis alone kills about 2 million people per year, mostly in sub-Saharan Africa. Antibiotics are used to treat bacterial infections and are also used in farming, making antibiotic resistance a growing problem. In industry, bacteria are important in sewage treatment and the breakdown of oil spills, the production of cheese and yogurt through fermentation, the recovery of gold, palladium, copper and other metals in the mining sector, as well as in biotechnology, and the manufacture of antibiotics and other chemicals.
Once regarded as plants constituting the class Schizomycetes (“fission fungi”), bacteria are now classified as prokaryotes. Unlike cells of animals and other eukaryotes, bacterial cells do not contain a nucleus and rarely harbour membrane-bound organelles. Although the term bacteria traditionally included all prokaryotes, the scientific classification changed after the discovery in the 1990s that prokaryotes consist of two very different groups of organisms that evolved from an ancient common ancestor. These evolutionary domains are called Bacteria and Archaea.
The ancestors of modern bacteria were unicellular microorganisms that were the first forms of life to appear on Earth, about 4 billion years ago. For about 3 billion years, most organisms were microscopic, and bacteria and archaea were the dominant forms of life. Although bacterial fossils exist, such as stromatolites, their lack of distinctive morphology prevents them from being used to examine the history of bacterial evolution, or to date the time of origin of a particular bacterial species. However, gene sequences can be used to reconstruct the bacterial phylogeny, and these studies indicate that bacteria diverged first from the archaeal/eukaryotic lineage. The most recent common ancestor of bacteria and archaea was probably a hyperthermophile that lived about 2.5 billion–3.2 billion years ago. The earliest life on land may have been bacteria some 3.22 billion years ago.
Bacteria were also involved in the second great evolutionary divergence, that of the archaea and eukaryotes. Here, eukaryotes resulted from the entering of ancient bacteria into endosymbiotic associations with the ancestors of eukaryotic cells, which were themselves possibly related to the Archaea. This involved the engulfment by proto-eukaryotic cells of alphaproteobacterial symbionts to form either mitochondria or hydrogenosomes, which are still found in all known Eukarya (sometimes in highly reduced form, e.g. in ancient “amitochondrial” protozoa). Later, some eukaryotes that already contained mitochondria also engulfed cyanobacteria-like organisms, leading to the formation of chloroplasts in algae and plants. This is known as primary endosymbiosis.
Bacteria display a wide diversity of shapes and sizes, called morphologies. Bacterial cells are about one-tenth the size of eukaryotic cells and are typically 0.5–5.0 micrometres in length. However, a few species are visible to the unaided eye—for example, Thiomargarita namibiensis is up to half a millimetre long and Epulopiscium fishelsoni reaches 0.7 mm. Among the smallest bacteria are members of the genus Mycoplasma, which measure only 0.3 micrometres, as small as the largest viruses. Some bacteria may be even smaller, but these ultramicrobacteria are not well-studied.
Most bacterial species are either spherical, called cocci (singular coccus, from Greek kókkos, grain, seed), or rod-shaped, called bacilli (sing. bacillus, from Latin baculus, stick). Some bacteria, called vibrio, are shaped like slightly curved rods or comma-shaped; others can be spiral-shaped, called spirilla, or tightly coiled, called spirochaetes. A small number of other unusual shapes have been described, such as star-shaped bacteria. This wide variety of shapes is determined by the bacterial cell wall and cytoskeleton, and is important because it can influence the ability of bacteria to acquire nutrients, attach to surfaces, swim through liquids and escape predators.
Many bacterial species exist simply as single cells, others associate in characteristic patterns: Neisseria form diploids (pairs), Streptococcus form chains, and Staphylococcus group together in “bunch of grapes” clusters. Bacteria can also group to form larger multicellular structures, such as the elongated filaments of Actinobacteria, the aggregates of Myxobacteria, and the complex hyphae of Streptomyces. These multicellular structures are often only seen in certain conditions. For example, when starved of amino acids, Myxobacteria detect surrounding cells in a process known as quorum sensing, migrate towards each other, and aggregate to form fruiting bodies up to 500 micrometres long and containing approximately 100,000 bacterial cells. In these fruiting bodies, the bacteria perform separate tasks; for example, about one in ten cells migrate to the top of a fruiting body and differentiate into a specialised dormant state called a myxospore, which is more resistant to drying and other adverse environmental conditions.
Bacteria often attach to surfaces and form dense aggregations called biofilms, and larger formations known as microbial mats. These biofilms and mats can range from a few micrometres in thickness to up to half a metre in depth, and may contain multiple species of bacteria, protists and archaea. Bacteria living in biofilms display a complex arrangement of cells and extracellular components, forming secondary structures, such as microcolonies, through which there are networks of channels to enable better diffusion of nutrients. In natural environments, such as soil or the surfaces of plants, the majority of bacteria are bound to surfaces in biofilms. Biofilms are also important in medicine, as these structures are often present during chronic bacterial infections or in infections of implanted medical devices, and bacteria protected within biofilms are much harder to kill than individual isolated bacteria.
4.6.1 Intracellular Structure
The bacterial cell is surrounded by a cell membrane, which is made primarily of phospholipids. This membrane encloses the contents of the cell and acts as a barrier to hold nutrients, proteins and other essential components of the cytoplasm within the cell. Unlike eukaryotic cells, bacteria usually lack large membrane-bound structures in their cytoplasm such as a nucleus, mitochondria, chloroplasts and the other organelles present in eukaryotic cells. However, some bacteria have protein-bound organelles in the cytoplasm which compartmentalize aspects of bacterial metabolism, such as the carboxysome. Additionally, bacteria have a multi-component cytoskeleton to control the localisation of proteins and nucleic acids within the cell, and to manage the process of cell division.
Many important biochemical reactions, such as energy generation, occur due to concentration gradients across membranes, creating a potential difference analogous to a battery. The general lack of internal membranes in bacteria means these reactions, such as electron transport, occur across the cell membrane between the cytoplasm and the outside of the cell or periplasm. However, in many photosynthetic bacteria the plasma membrane is highly folded and fills most of the cell with layers of light-gathering membrane. These light-gathering complexes may even form lipid-enclosed structures called chlorosomes in green sulfur bacteria.
Bacteria do not have a membrane-bound nucleus, and their genetic material is typically a single circular bacterial chromosome of DNA located in the cytoplasm in an irregularly shaped body called the nucleoid. The nucleoid contains the chromosome with its associated proteins and RNA. Like all other organisms, bacteria contain ribosomes for the production of proteins, but the structure of the bacterial ribosome is different from that of eukaryotes and Archaea.
Some bacteria produce intracellular nutrient storage granules, such as glycogen, polyphosphate, sulfur or polyhydroxyalkanoates. Bacteria such as the photosynthetic cyanobacteria, produce internal gas vacuoles, which they use to regulate their buoyancy, allowing them to move up or down into water layers with different light intensities and nutrient levels.
4.6.2 Extracellular Structures
Around the outside of the cell membrane is the cell wall. Bacterial cell walls are made of peptidoglycan (also called murein), which is made from polysaccharide chains cross-linked by peptides containing D-amino acids. Bacterial cell walls are different from the cell walls of plants and fungi, which are made of cellulose and chitin, respectively. The cell wall of bacteria is also distinct from that of Archaea, which do not contain peptidoglycan. The cell wall is essential to the survival of many bacteria, and the antibiotic penicillin (produced by a fungus called Penicillium) is able to kill bacteria by inhibiting a step in the synthesis of peptidoglycan.
There are broadly speaking two different types of cell wall in bacteria, that classify bacteria into Gram-positive bacteria and Gram-negative bacteria. The names originate from the reaction of cells to the Gram stain, a long-standing test for the classification of bacterial species.
Gram-positive bacteria possess a thick cell wall containing many layers of peptidoglycan and teichoic acids. In contrast, Gram-negative bacteria have a relatively thin cell wall consisting of a few layers of peptidoglycan surrounded by a second lipid membrane containing lipopolysaccharides and lipoproteins. Most bacteria have the Gram-negative cell wall, and only the Firmicutes and Actinobacteria (previously known as the low G+C and high G+C Gram-positive bacteria, respectively) have the alternative Gram-positive arrangement. These differences in structure can produce differences in antibiotic susceptibility; for instance, vancomycin can kill only Gram-positive bacteria and is ineffective against Gram-negative pathogens, such as Haemophilus influenzae or Pseudomonas aeruginosa. Some bacteria have cell wall structures that are neither classically Gram-positive or Gram-negative. This includes clinically important bacteria such as Mycobacteria which have a thick peptidoglycan cell wall like a Gram-positive bacterium, but also a second outer layer of lipids.
In many bacteria, an S-layer of rigidly arrayed protein molecules covers the outside of the cell. This layer provides chemical and physical protection for the cell surface and can act as a macromolecular diffusion barrier. S-layers have diverse but mostly poorly understood functions, but are known to act as virulence factors in Campylobacter and contain surface enzymes in Bacillus stearothermophilus.
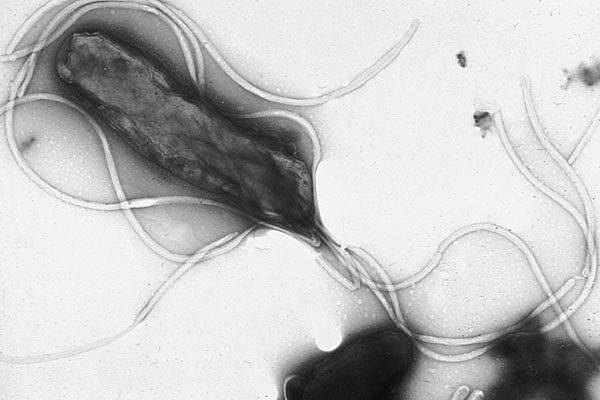
Figure 4.12: Helicobacter pylori electron micrograph, showing multiple flagella on the cell surface
Flagella are rigid protein structures, about 20 nanometres in diameter and up to 20 micrometres in length, that are used for motility. Flagella are driven by the energy released by the transfer of ions down an electrochemical gradient across the cell membrane.
The bacterial flagellum is made up of the protein flagellin. Its shape is a 20-nanometer-thick hollow tube. It is helical and has a sharp bend just outside the outer membrane; this “hook” allows the axis of the helix to point directly away from the cell. A shaft runs between the hook and the basal body, passing through protein rings in the cell’s membrane that act as bearings. Gram-positive organisms have two of these basal body rings, one in the peptidoglycan layer and one in the plasma membrane. Gram-negative organisms have four such rings: the L ring associates with the lipopolysaccharides, the P ring associates with peptidoglycan layer, the M ring is embedded in the plasma membrane, and the S ring is directly attached to the plasma membrane. The filament ends with a capping protein.
Figure 4.13: Gram-negative bacterial flagellum. A flagellum is a long, slender projection from the cell body, whose function is to propel a unicellular or small multicellular organism. The depicted type of flagellum is found in bacteria such as E. coli and Salmonella, and rotates like a propeller when the bacterium swims. The bacterial movement can be divided into 2 kinds: run, resulting from a counterclockwise rotation of the flagellum, and tumbling, from a clockwise rotation of the flagellum.
The flagellar filament is the long, helical screw that propels the bacterium when rotated by the motor, through the hook. In most bacteria that have been studied, including the Gram-negative Escherichia coli, Salmonella typhimurium, Caulobacter crescentus, and Vibrio alginolyticus, the filament is made up of 11 protofilaments approximately parallel to the filament axis. Each protofilament is a series of tandem protein chains. However, Campylobacter jejuni has seven protofilaments.
The basal body has several traits in common with some types of secretory pores, such as the hollow, rod-like “plug” in their centers extending out through the plasma membrane. The similarities between bacterial flagella and bacterial secretory system structures and proteins provide scientific evidence supporting the theory that bacterial flagella evolved from the type-three secretion system.
The bacterial flagellum is driven by a rotary engine (Mot complex) made up of protein, located at the flagellum’s anchor point on the inner cell membrane. The engine is powered by proton motive force, i.e., by the flow of protons (hydrogen ions) across the bacterial cell membrane due to a concentration gradient set up by the cell’s metabolism (Vibrio species have two kinds of flagella, lateral and polar, and some are driven by a sodium ion pump rather than a proton pump). The rotor transports protons across the membrane, and is turned in the process. The rotor alone can operate at 6,000 to 17,000 rpm, but with the flagellar filament attached usually only reaches 200 to 1000 rpm. The direction of rotation can be changed by the flagellar motor switch almost instantaneously, caused by a slight change in the position of a protein, FliG, in the rotor. The flagellum is highly energy efficient and uses very little energy.[unreliable source?] The exact mechanism for torque generation is still poorly understood. Because the flagellar motor has no on-off switch, the protein epsE is used as a mechanical clutch to disengage the motor from the rotor, thus stopping the flagellum and allowing the bacterium to remain in one place.
The cylindrical shape of flagella is suited to locomotion of microscopic organisms; these organisms operate at a low Reynolds number, where the viscosity of the surrounding water is much more important than its mass or inertia.
The rotational speed of flagella varies in response to the intensity of the proton motive force, thereby permitting certain forms of speed control, and also permitting some types of bacteria to attain remarkable speeds in proportion to their size; some achieve roughly 60 cell lengths per second. At such a speed, a bacterium would take about 245 days to cover 1 km; although that may seem slow, the perspective changes when the concept of scale is introduced. In comparison to macroscopic life forms, it is very fast indeed when expressed in terms of number of body lengths per second. A cheetah, for example, only achieves about 25 body lengths per second.
Through use of their flagella, E. coli is able to move rapidly towards attractants and away from repellents, by means of a biased random walk, with ‘runs’ and ‘tumbles’ brought about by rotating its flagellum counterclockwise and clockwise, respectively. The two directions of rotation are not identical (with respect to flagellum movement) and are selected by a molecular switch.
During flagellar assembly, components of the flagellum pass through the hollow cores of the basal body and the nascent filament. During assembly, protein components are added at the flagellar tip rather than at the base. In vitro, flagellar filaments assemble spontaneously in a solution containing purified flagellin as the sole protein.
At least 10 protein components of the bacterial flagellum share homologous proteins with the type three secretion system (T3SS) found in many gram-negative bacteria, hence one likely evolved from the other. Because the T3SS has a similar number of components as a flagellar apparatus (about 25 proteins), which one evolved first is difficult to determine. However, the flagellar system appears to involve more proteins overall, including various regulators and chaperones, hence it has been argued that flagella evolved from a T3SS. However, it has also been suggested that the flagellum may have evolved first or the two structures evolved in parallel. Early single-cell organisms’ need for motility (mobility) support that the more mobile flagella would be selected by evolution first, but the T3SS evolving from the flagellum can be seen as ‘reductive evolution’, and receives no topological support from the phylogenetic trees. The hypothesis that the two structures evolved separately from a common ancestor accounts for the protein similarities between the two structures, as well as their functional diversity.
Different species of bacteria have different numbers and arrangements of flagella.
- Monotrichous bacteria have a single flagellum (e.g., Vibrio cholerae).
- Lophotrichous bacteria have multiple flagella located at the same spot on the bacterial surfaces which act in concert to drive the bacteria in a single direction. In many cases, the bases of multiple flagella are surrounded by a specialized region of the cell membrane, called the polar organelle.[citation needed]
- Amphitrichous bacteria have a single flagellum on each of two opposite ends (only one flagellum operates at a time, allowing the bacterium to reverse course rapidly by switching which flagellum is active).
- Peritrichous bacteria have flagella projecting in all directions (e.g., E. coli).
In certain large forms of Selenomonas, more than 30 individual flagella are organized outside the cell body, helically twining about each other to form a thick structure (easily visible with the light microscope) called a “fascicle”.
Spirochetes, in contrast, have flagella called endoflagella arising from opposite poles of the cell, and are located within the periplasmic space as shown by breaking the outer-membrane and also by electron cryotomography microscopy. The rotation of the filaments relative to the cell body causes the entire bacterium to move forward in a corkscrew-like motion, even through material viscous enough to prevent the passage of normally flagellated bacteria.
Counterclockwise rotation of a monotrichous polar flagellum pushes the cell forward with the flagellum trailing behind, much like a corkscrew moving inside cork. Indeed, water on the microscopic scale is highly viscous, very different from our daily experience of water.
Flagella are left-handed helices, and bundle and rotate together only when rotating counterclockwise. When some of the rotors reverse direction, the flagella unwind and the cell starts “tumbling”. Even if all flagella would rotate clockwise, they likely will not form a bundle, due to geometrical, as well as hydrodynamic reasons. Such “tumbling” may happen occasionally, leading to the cell seemingly thrashing about in place, resulting in the reorientation of the cell. The clockwise rotation of a flagellum is suppressed by chemical compounds favorable to the cell (e.g. food), but the motor is highly adaptive to this. Therefore, when moving in a favorable direction, the concentration of the chemical attractant increases and “tumbles” are continually suppressed; however, when the cell’s direction of motion is unfavorable (e.g., away from a chemical attractant), tumbles are no longer suppressed and occur much more often, with the chance that the cell will be thus reoriented in the correct direction.
In some Vibrio spp. (particularly Vibrio parahaemolyticus) and related proteobacteria such as Aeromonas, two flagellar systems co-exist, using different sets of genes and different ion gradients for energy. The polar flagella are constitutively expressed and provide motility in bulk fluid, while the lateral flagella are expressed when the polar flagella meet too much resistance to turn. These provide swarming motility on surfaces or in viscous fluids.
Fimbriae (sometimes called “attachment pili”) are fine filaments of protein, usually 2–10 nanometres in diameter and up to several micrometres in length. They are distributed over the surface of the cell, and resemble fine hairs when seen under the electron microscope. Fimbriae are believed to be involved in attachment to solid surfaces or to other cells, and are essential for the virulence of some bacterial pathogens. Pili (sing. pilus) are cellular appendages, slightly larger than fimbriae, that can transfer genetic material between bacterial cells in a process called conjugation where they are called conjugation pili or sex pili (see bacterial genetics, below). They can also generate movement where they are called type IV pili.
Glycocalyx is produced by many bacteria to surround their cells, and varies in structural complexity: ranging from a disorganised slime layer of extracellular polymeric substances to a highly structured capsule. These structures can protect cells from engulfment by eukaryotic cells such as macrophages (part of the human immune system). They can also act as antigens and be involved in cell recognition, as well as aiding attachment to surfaces and the formation of biofilms.
The assembly of these extracellular structures is dependent on bacterial secretion systems. These transfer proteins from the cytoplasm into the periplasm or into the environment around the cell. Many types of secretion systems are known and these structures are often essential for the virulence of pathogens, so are intensively studied.
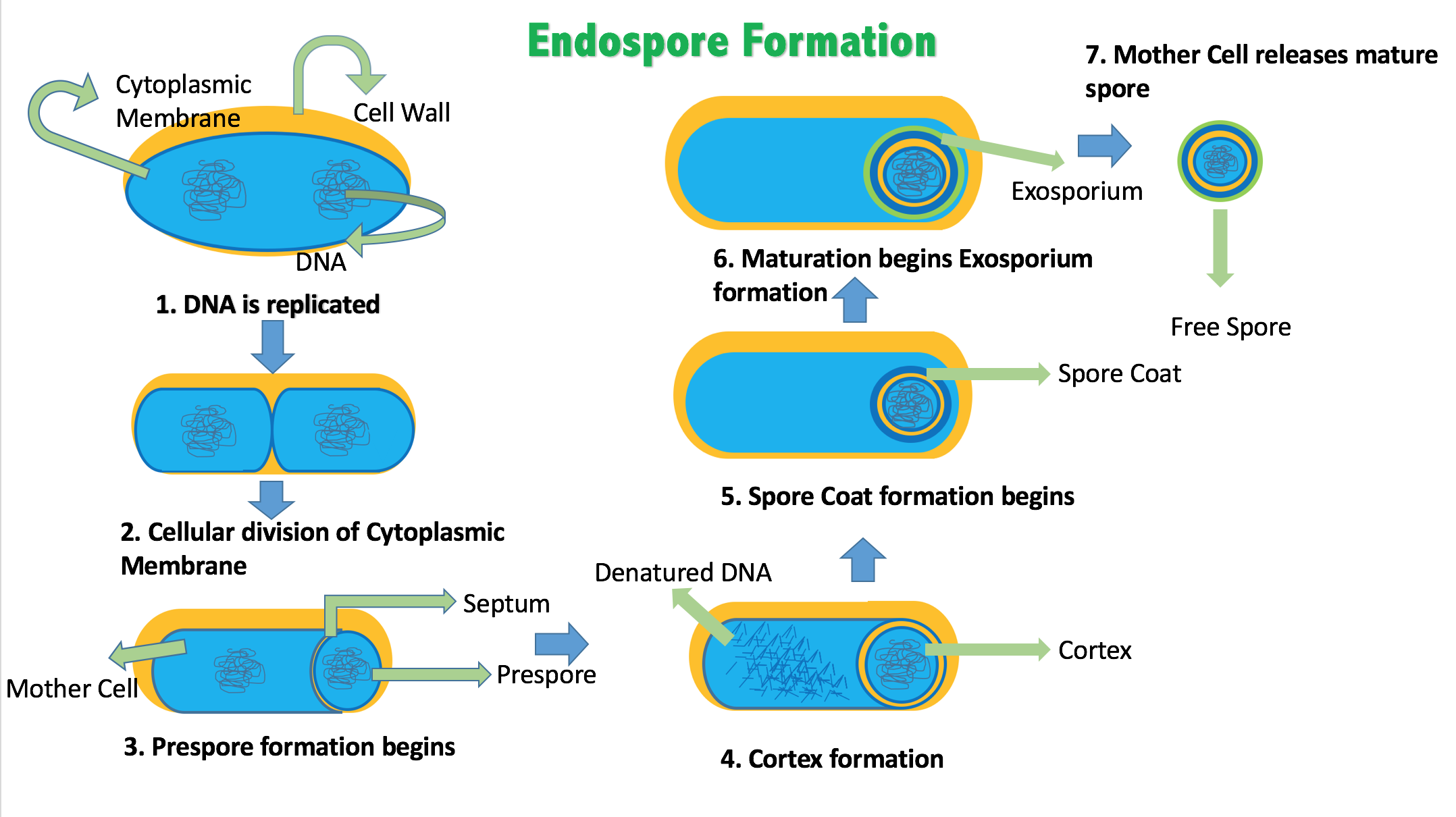
4.6.3 Endospores
Certain genera of Gram-positive bacteria, such as Bacillus, Clostridium, Sporohalobacter, Anaerobacter, and Heliobacterium, can form highly resistant, dormant structures called endospores. Endospores develop within the cytoplasm of the cell; generally a single endospore develops in each cell. Each endospore contains a core of DNA and ribosomes surrounded by a cortex layer and protected by a multilayer rigid coat composed of peptidoglycan and a variety of proteins.
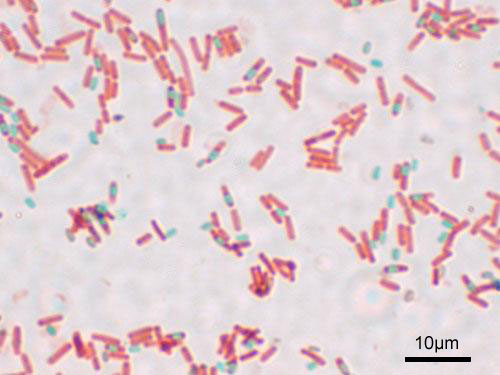
Figure 4.14: A stained preparation of the cell Bacillus subtilis showing endospores as green and the vegetative cell as red.
Endospores show no detectable metabolism and can survive extreme physical and chemical stresses, such as high levels of UV light, gamma radiation, detergents, disinfectants, heat, freezing, pressure, and desiccation. In this dormant state, these organisms may remain viable for millions of years, and endospores even allow bacteria to survive exposure to the vacuum and radiation in space, possibly bacteria could be distributed throughout the Universe by space dust, meteoroids, asteroids, comets, planetoids or via directed panspermia. Endospore-forming bacteria can also cause disease: for example, anthrax can be contracted by the inhalation of Bacillus anthracis endospores, and contamination of deep puncture wounds with Clostridium tetani endospores causes tetanus.
4.6.4 Metabolism
Bacteria exhibit an extremely wide variety of metabolic types. The distribution of metabolic traits within a group of bacteria has traditionally been used to define their taxonomy, but these traits often do not correspond with modern genetic classifications. Bacterial metabolism is classified into nutritional groups on the basis of three major criteria: the source of energy, the electron donors used, and the source of carbon used for growth.
Bacteria either derive energy from light using photosynthesis (called phototrophy), or by breaking down chemical compounds using oxidation (called chemotrophy). Chemotrophs use chemical compounds as a source of energy by transferring electrons from a given electron donor to a terminal electron acceptor in a redox reaction. This reaction releases energy that can be used to drive metabolism. Chemotrophs are further divided by the types of compounds they use to transfer electrons. Bacteria that use inorganic compounds such as hydrogen, carbon monoxide, or ammonia as sources of electrons are called lithotrophs, while those that use organic compounds are called organotrophs. The compounds used to receive electrons are also used to classify bacteria: aerobic organisms use oxygen as the terminal electron acceptor, while anaerobic organisms use other compounds such as nitrate, sulfate, or carbon dioxide.
Many bacteria get their carbon from other organic carbon, called heterotrophy. Others such as cyanobacteria and some purple bacteria are autotrophic, meaning that they obtain cellular carbon by fixing carbon dioxide. In unusual circumstances, the gas methane can be used by methanotrophic bacteria as both a source of electrons and a substrate for carbon anabolism.
| Nutritional Type | Source of Energy | Source of Carbon | Examples |
|---|---|---|---|
| Phototrophs | Sunlight | Organic compounds (photoheterotrophs) or carbon fixation (photoautotrophs) | Cyanobacteria, Green sulfur bacteria, Chloroflexi, or Purple bacteria |
| Lithotrophs | Inorganic compounds | Organic compounds (lithoheterotrophs) or carbon fixation (lithoautotrophs) | Thermodesulfobacteria, Hydrogenophilaceae, or Nitrospirae |
| Organotrophs | Organic compounds | Organic compounds (chemoheterotrophs) or carbon fixation (chemoautotrophs) | Bacillus, Clostridium or Enterobacteriaceae |
In many ways, bacterial metabolism provides traits that are useful for ecological stability and for human society. One example is that some bacteria have the ability to fix nitrogen gas using the enzyme nitrogenase. This environmentally important trait can be found in bacteria of most metabolic types listed above. This leads to the ecologically important processes of denitrification, sulfate reduction, and acetogenesis, respectively. Bacterial metabolic processes are also important in biological responses to pollution; for example, sulfate-reducing bacteria are largely responsible for the production of the highly toxic forms of mercury (methyl- and dimethylmercury) in the environment. Non-respiratory anaerobes use fermentation to generate energy and reducing power, secreting metabolic by-products (such as ethanol in brewing) as waste. Facultative anaerobes can switch between fermentation and different terminal electron acceptors depending on the environmental conditions in which they find themselves.
4.6.5 Growth And Reproduction
Many bacteria reproduce through binary fission, which is compared to mitosis and meiosis in this image.
Unlike in multicellular organisms, increases in cell size (cell growth) and reproduction by cell division are tightly linked in unicellular organisms. Bacteria grow to a fixed size and then reproduce through binary fission, a form of asexual reproduction. Under optimal conditions, bacteria can grow and divide extremely rapidly, and bacterial populations can double as quickly as every 9.8 minutes. In cell division, two identical clone daughter cells are produced. Some bacteria, while still reproducing asexually, form more complex reproductive structures that help disperse the newly formed daughter cells. Examples include fruiting body formation by Myxobacteria and aerial hyphae formation by Streptomyces, or budding. Budding involves a cell forming a protrusion that breaks away and produces a daughter cell.
In the laboratory, bacteria are usually grown using solid or liquid media. Solid growth media, such as agar plates, are used to isolate pure cultures of a bacterial strain. However, liquid growth media are used when the measurement of growth or large volumes of cells are required. Growth in stirred liquid media occurs as an even cell suspension, making the cultures easy to divide and transfer, although isolating single bacteria from liquid media is difficult. The use of selective media (media with specific nutrients added or deficient, or with antibiotics added) can help identify specific organisms.
Most laboratory techniques for growing bacteria use high levels of nutrients to produce large amounts of cells cheaply and quickly. However, in natural environments, nutrients are limited, meaning that bacteria cannot continue to reproduce indefinitely. This nutrient limitation has led the evolution of different growth strategies (see r/K selection theory). Some organisms can grow extremely rapidly when nutrients become available, such as the formation of algal (and cyanobacterial) blooms that often occur in lakes during the summer. Other organisms have adaptations to harsh environments, such as the production of multiple antibiotics by Streptomyces that inhibit the growth of competing microorganisms. In nature, many organisms live in communities (e.g., biofilms) that may allow for increased supply of nutrients and protection from environmental stresses. These relationships can be essential for growth of a particular organism or group of organisms (syntrophy).
Bacterial growth follows four phases. When a population of bacteria first enter a high-nutrient environment that allows growth, the cells need to adapt to their new environment. The first phase of growth is the lag phase, a period of slow growth when the cells are adapting to the high-nutrient environment and preparing for fast growth. The lag phase has high biosynthesis rates, as proteins necessary for rapid growth are produced. The second phase of growth is the logarithmic phase, also known as the exponential phase. The log phase is marked by rapid exponential growth. The rate at which cells grow during this phase is known as the growth rate (k), and the time it takes the cells to double is known as the generation time (g). During log phase, nutrients are metabolised at maximum speed until one of the nutrients is depleted and starts limiting growth. The third phase of growth is the stationary phase and is caused by depleted nutrients. The cells reduce their metabolic activity and consume non-essential cellular proteins. The stationary phase is a transition from rapid growth to a stress response state and there is increased expression of genes involved in DNA repair, antioxidant metabolism and nutrient transport. The final phase is the death phase where the bacteria run out of nutrients and die.
4.6.6 Bacterial Genetics
Most bacteria have a single circular chromosome that can range in size from only 160,000 base pairs in the endosymbiotic bacteria Carsonella ruddii, to 12,200,000 base pairs (12.2 Mbp) in the soil-dwelling bacteria Sorangium cellulosum. There are many exceptions to this, for example some Streptomyces and Borrelia species contain a single linear chromosome, while some Vibrio species contain more than one chromosome. Bacteria can also contain plasmids, small extra-chromosomal molecules of DNA that may contain genes for various useful functions such as antibiotic resistance, metabolic capabilities, or various virulence factors.
Bacteria genomes usually encode a few hundred to a few thousand genes. The genes in bacterial genomes are usually a single continuous stretch of DNA and although several different types of introns do exist in bacteria, these are much rarer than in eukaryotes.
Bacteria, as asexual organisms, inherit an identical copy of the parent’s genomes and are clonal. However, all bacteria can evolve by selection on changes to their genetic material DNA caused by genetic recombination or mutations. Mutations come from errors made during the replication of DNA or from exposure to mutagens. Mutation rates vary widely among different species of bacteria and even among different clones of a single species of bacteria. Genetic changes in bacterial genomes come from either random mutation during replication or “stress-directed mutation”, where genes involved in a particular growth-limiting process have an increased mutation rate.
Some bacteria also transfer genetic material between cells. This can occur in three main ways. First, bacteria can take up exogenous DNA from their environment, in a process called transformation. Many bacteria can naturally take up DNA from the environment, while others must be chemically altered in order to induce them to take up DNA. The development of competence in nature is usually associated with stressful environmental conditions, and seems to be an adaptation for facilitating repair of DNA damage in recipient cells. The second way bacteria transfer genetic material is by transduction, when the integration of a bacteriophage introduces foreign DNA into the chromosome. Many types of bacteriophage exist, some simply infect and lyse their host bacteria, while others insert into the bacterial chromosome. Bacteria resist phage infection through restriction modification systems that degrade foreign DNA, and a system that uses CRISPR sequences to retain fragments of the genomes of phage that the bacteria have come into contact with in the past, which allows them to block virus replication through a form of RNA interference. The third method of gene transfer is conjugation, whereby DNA is transferred through direct cell contact. In ordinary circumstances, transduction, conjugation, and transformation involve transfer of DNA between individual bacteria of the same species, but occasionally transfer may occur between individuals of different bacterial species and this may have significant consequences, such as the transfer of antibiotic resistance. In such cases, gene acquisition from other bacteria or the environment is called horizontal gene transfer and may be common under natural conditions.
4.6.7 Movement Of Bacteria
Many bacteria are motile (able to move themselves) and do so using a variety of mechanisms. The best studied of these are flagella, long filaments that are turned by a motor at the base to generate propeller-like movement. The bacterial flagellum is made of about 20 proteins, with approximately another 30 proteins required for its regulation and assembly. The flagellum is a rotating structure driven by a reversible motor at the base that uses the electrochemical gradient across the membrane for power.
Bacteria can use flagella in different ways to generate different kinds of movement. Many bacteria (such as E. coli) have two distinct modes of movement: forward movement (swimming) and tumbling. The tumbling allows them to reorient and makes their movement a three-dimensional random walk. Bacterial species differ in the number and arrangement of flagella on their surface; some have a single flagellum (monotrichous), a flagellum at each end (amphitrichous), clusters of flagella at the poles of the cell (lophotrichous), while others have flagella distributed over the entire surface of the cell (peritrichous). The flagella of a unique group of bacteria, the spirochaetes, are found between two membranes in the periplasmic space. They have a distinctive helical body that twists about as it moves.
Two other types of bacterial motion are called twitching motility that relies on a structure called the type IV pilus, and gliding motility, that uses other mechanisms. In twitching motility, the rod-like pilus extends out from the cell, binds some substrate, and then retracts, pulling the cell forward.
Motile bacteria are attracted or repelled by certain stimuli in behaviours called taxes: these include chemotaxis, phototaxis, energy taxis, and magnetotaxis. In one peculiar group, the myxobacteria, individual bacteria move together to form waves of cells that then differentiate to form fruiting bodies containing spores. The myxobacteria move only when on solid surfaces, unlike E. coli, which is motile in liquid or solid media.
Several Listeria and Shigella species move inside host cells by usurping the cytoskeleton, which is normally used to move organelles inside the cell. By promoting actin polymerisation at one pole of their cells, they can form a kind of tail that pushes them through the host cell’s cytoplasm.
4.6.8 Bacterial Communication
Bacteria often function as multicellular aggregates known as biofilms, exchanging a variety of molecular signals for inter-cell communication, and engaging in coordinated multicellular behaviour.
The communal benefits of multicellular cooperation include a cellular division of labour, accessing resources that cannot effectively be used by single cells, collectively defending against antagonists, and optimising population survival by differentiating into distinct cell types. For example, bacteria in biofilms can have more than 500 times increased resistance to antibacterial agents than individual “planktonic” bacteria of the same species.
One type of inter-cellular communication by a molecular signal is called quorum sensing, which serves the purpose of determining whether there is a local population density that is sufficiently high that it is productive to invest in processes that are only successful if large numbers of similar organisms behave similarly, as in excreting digestive enzymes or emitting light.
Quorum sensing allows bacteria to coordinate gene expression, and enables them to produce, release and detect autoinducers or pheromones which accumulate with the growth in cell population.
4.6.9 Classification And identification
Classification seeks to describe the diversity of bacterial species by naming and grouping organisms based on similarities. Bacteria can be classified on the basis of cell structure, cellular metabolism or on differences in cell components, such as DNA, fatty acids, pigments, antigens and quinones. While these schemes allowed the identification and classification of bacterial strains, it was unclear whether these differences represented variation between distinct species or between strains of the same species. This uncertainty was due to the lack of distinctive structures in most bacteria, as well as lateral gene transfer between unrelated species. Due to lateral gene transfer, some closely related bacteria can have very different morphologies and metabolisms. To overcome this uncertainty, modern bacterial classification emphasises molecular systematics, using genetic techniques such as guanine cytosine ratio determination, genome-genome hybridisation, as well as sequencing genes that have not undergone extensive lateral gene transfer, such as the rRNA gene. Classification of bacteria is determined by publication in the International Journal of Systematic Bacteriology, and Bergey’s Manual of Systematic Bacteriology. The International Committee on Systematic Bacteriology (ICSB) maintains international rules for the naming of bacteria and taxonomic categories and for the ranking of them in the International Code of Nomenclature of Bacteria.
The term “bacteria” was traditionally applied to all microscopic, single-cell prokaryotes. However, molecular systematics showed prokaryotic life to consist of two separate domains, originally called Eubacteria and Archaebacteria, but now called Bacteria and Archaea that evolved independently from an ancient common ancestor. The archaea and eukaryotes are more closely related to each other than either is to the bacteria. These two domains, along with Eukarya, are the basis of the three-domain system, which is currently the most widely used classification system in microbiology. However, due to the relatively recent introduction of molecular systematics and a rapid increase in the number of genome sequences that are available, bacterial classification remains a changing and expanding field. For example, Cavalier-Smith argued that the Archaea and Eukaryotes evolved from Gram-positive bacteria.
The identification of bacteria in the laboratory is particularly relevant in medicine, where the correct treatment is determined by the bacterial species causing an infection. Consequently, the need to identify human pathogens was a major impetus for the development of techniques to identify bacteria.
The Gram stain, developed in 1884 by Hans Christian Gram, characterises bacteria based on the structural characteristics of their cell walls. The thick layers of peptidoglycan in the “Gram-positive” cell wall stain purple, while the thin “Gram-negative” cell wall appears pink. By combining morphology and Gram-staining, most bacteria can be classified as belonging to one of four groups (Gram-positive cocci, Gram-positive bacilli, Gram-negative cocci and Gram-negative bacilli). Some organisms are best identified by stains other than the Gram stain, particularly mycobacteria or Nocardia, which show acid-fastness on Ziehl–Neelsen or similar stains. Other organisms may need to be identified by their growth in special media, or by other techniques, such as serology.
Culture techniques are designed to promote the growth and identify particular bacteria, while restricting the growth of the other bacteria in the sample. Often these techniques are designed for specific specimens; for example, a sputum sample will be treated to identify organisms that cause pneumonia, while stool specimens are cultured on selective media to identify organisms that cause diarrhoea, while preventing growth of non-pathogenic bacteria. Specimens that are normally sterile, such as blood, urine or spinal fluid, are cultured under conditions designed to grow all possible organisms. Once a pathogenic organism has been isolated, it can be further characterised by its morphology, growth patterns (such as aerobic or anaerobic growth), patterns of hemolysis, and staining.
As with bacterial classification, identification of bacteria is increasingly using molecular methods. Diagnostics using DNA-based tools, such as polymerase chain reaction, are increasingly popular due to their specificity and speed, compared to culture-based methods. These methods also allow the detection and identification of “viable but nonculturable” cells that are metabolically active but non-dividing. However, even using these improved methods, the total number of bacterial species is not known and cannot even be estimated with any certainty. Following present classification, there are a little less than 9,300 known species of prokaryotes, which includes bacteria and archaea; but attempts to estimate the true number of bacterial diversity have ranged from 107 to 109 total species—and even these diverse estimates may be off by many orders of magnitude.
4.6.10 Interactions With Other Organisms
Despite their apparent simplicity, bacteria can form complex associations with other organisms. These symbiotic associations can be divided into parasitism, mutualism and commensalism. Due to their small size, commensal bacteria are ubiquitous and grow on animals and plants exactly as they will grow on any other surface. However, their growth can be increased by warmth and sweat, and large populations of these organisms in humans are the cause of body odour.
Some species of bacteria kill and then consume other microorganisms, these species are called predatory bacteria. These include organisms such as Myxococcus xanthus, which forms swarms of cells that kill and digest any bacteria they encounter. Other bacterial predators either attach to their prey in order to digest them and absorb nutrients, such as Vampirovibrio chlorellavorus, or invade another cell and multiply inside the cytosol, such as Daptobacter. These predatory bacteria are thought to have evolved from saprophages that consumed dead microorganisms, through adaptations that allowed them to entrap and kill other organisms.
Certain bacteria form close spatial associations that are essential for their survival. One such mutualistic association, called interspecies hydrogen transfer, occurs between clusters of anaerobic bacteria that consume organic acids, such as butyric acid or propionic acid, and produce hydrogen, and methanogenic Archaea that consume hydrogen. The bacteria in this association are unable to consume the organic acids as this reaction produces hydrogen that accumulates in their surroundings. Only the intimate association with the hydrogen-consuming Archaea keeps the hydrogen concentration low enough to allow the bacteria to grow.
In soil, microorganisms that reside in the rhizosphere (a zone that includes the root surface and the soil that adheres to the root after gentle shaking) carry out nitrogen fixation, converting nitrogen gas to nitrogenous compounds. This serves to provide an easily absorbable form of nitrogen for many plants, which cannot fix nitrogen themselves. Many other bacteria are found as symbionts in humans and other organisms. For example, the presence of over 1,000 bacterial species in the normal human gut flora of the intestines can contribute to gut immunity, synthesise vitamins, such as folic acid, vitamin K and biotin, convert sugars to lactic acid (see Lactobacillus), as well as fermenting complex undigestible carbohydrates. The presence of this gut flora also inhibits the growth of potentially pathogenic bacteria (usually through competitive exclusion) and these beneficial bacteria are consequently sold as probiotic dietary supplements.
4.6.11 Bacteria As Pathogens
If bacteria form a parasitic association with other organisms, they are classed as pathogens. Pathogenic bacteria are a major cause of human death and disease and cause infections such as tetanus (Caused by Clostridium tetani), typhoid fever, diphtheria, syphilis, cholera, foodborne illness, leprosy (caused by Micobacterium leprae) and tuberculosis (Caused by Mycobacterium tuberculosis). A pathogenic cause for a known medical disease may only be discovered many years after, as was the case with Helicobacter pylori and peptic ulcer disease. Bacterial diseases are also important in agriculture, with bacteria causing leaf spot, fire blight and wilts in plants, as well as Johne’s disease, mastitis, salmonella and anthrax in farm animals.
Each species of pathogen has a characteristic spectrum of interactions with its human hosts. Some organisms, such as Staphylococcus or Streptococcus, can cause skin infections, pneumonia, meningitis and even overwhelming sepsis, a systemic inflammatory response producing shock, massive vasodilation and death. Yet these organisms are also part of the normal human flora and usually exist on the skin or in the nose without causing any disease at all. Other organisms invariably cause disease in humans, such as the Rickettsia, which are obligate intracellular parasites able to grow and reproduce only within the cells of other organisms. One species of Rickettsia causes typhus, while another causes Rocky Mountain spotted fever. Chlamydia, another phylum of obligate intracellular parasites, contains species that can cause pneumonia, or urinary tract infection and may be involved in coronary heart disease. Finally, some species, such as Pseudomonas aeruginosa, Burkholderia cenocepacia, and Mycobacterium avium, are opportunistic pathogens and cause disease mainly in people suffering from immunosuppression or cystic fibrosis.
Bacterial infections may be treated with antibiotics, which are classified as bacteriocidal if they kill bacteria, or bacteriostatic if they just prevent bacterial growth. There are many types of antibiotics and each class inhibits a process that is different in the pathogen from that found in the host. An example of how antibiotics produce selective toxicity are chloramphenicol and puromycin, which inhibit the bacterial ribosome, but not the structurally different eukaryotic ribosome. Antibiotics are used both in treating human disease and in intensive farming to promote animal growth, where they may be contributing to the rapid development of antibiotic resistance in bacterial populations. Infections can be prevented by antiseptic measures such as sterilising the skin prior to piercing it with the needle of a syringe, and by proper care of indwelling catheters. Surgical and dental instruments are also sterilised to prevent contamination by bacteria. Disinfectants such as bleach are used to kill bacteria or other pathogens on surfaces to prevent contamination and further reduce the risk of infection.
4.6.12 Significance In Technology And Industry
Bacteria, often lactic acid bacteria, such as Lactobacillus and Lactococcus, in combination with yeasts and moulds, have been used for thousands of years in the preparation of fermented foods, such as cheese, pickles, soy sauce, sauerkraut, vinegar, wine and yogurt.
The ability of bacteria to degrade a variety of organic compounds is remarkable and has been used in waste processing and bioremediation. Bacteria capable of digesting the hydrocarbons in petroleum are often used to clean up oil spills. Fertiliser was added to some of the beaches in Prince William Sound in an attempt to promote the growth of these naturally occurring bacteria after the 1989 Exxon Valdez oil spill. These efforts were effective on beaches that were not too thickly covered in oil. Bacteria are also used for the bioremediation of industrial toxic wastes. In the chemical industry, bacteria are most important in the production of enantiomerically pure chemicals for use as pharmaceuticals or agrichemicals.
Bacteria can also be used in the place of pesticides in the biological pest control. This commonly involves Bacillus thuringiensis (also called BT), a Gram-positive, soil dwelling bacterium. Subspecies of this bacteria are used as a Lepidopteran-specific insecticides under trade names such as Dipel and Thuricide. Because of their specificity, these pesticides are regarded as environmentally friendly, with little or no effect on humans, wildlife, pollinators and most other beneficial insects.
Because of their ability to quickly grow and the relative ease with which they can be manipulated, bacteria are the workhorses for the fields of molecular biology, genetics and biochemistry. By making mutations in bacterial DNA and examining the resulting phenotypes, scientists can determine the function of genes, enzymes and metabolic pathways in bacteria, then apply this knowledge to more complex organisms. This aim of understanding the biochemistry of a cell reaches its most complex expression in the synthesis of huge amounts of enzyme kinetic and gene expression data into mathematical models of entire organisms. This is achievable in some well-studied bacteria, with models of Escherichia coli metabolism now being produced and tested. This understanding of bacterial metabolism and genetics allows the use of biotechnology to bioengineer bacteria for the production of therapeutic proteins, such as insulin, growth factors, or antibodies.
Because of their importance for research in general, samples of bacterial strains are isolated and preserved in Biological Resource Centers. This ensures the availability of the strain to scientists worldwide.
4.7 Archaea
The word archaea comes from the Ancient Greek ἀρχαῖα, meaning “ancient things”, as the first representatives of the domain Archaea were methanogens and it was assumed that their metabolism reflected Earth’s primitive atmosphere and the organisms’ antiquity, but as new habitats were studied, more organisms were discovered. Extreme halophilic and hyperthermophilic microbes were also included in Archaea. For a long time, archaea were seen as extremophiles that exist only in extreme habitats such as hot springs and salt lakes, but by the end of the 20th century, archaea had been identified in non-extreme environments as well. Today, they are known to be a large and diverse group of organisms abundantly distributed throughout nature. This new appreciation of the importance and ubiquity of archaea came from using polymerase chain reaction (PCR) to detect prokaryotes from environmental samples (such as water or soil) by multiplying their ribosomal genes. This allows the detection and identification of organisms that have not been cultured in the laboratory.
Archaeal cells have unique properties separating them from the other two domains, Bacteria and Eukaryota. Archaea are further divided into multiple recognized phyla. Classification is difficult because most have not been isolated in the laboratory and have been detected only by analysis of their nucleic acids in samples from their environment.
Archaea and bacteria are generally similar in size and shape, although a few archaea have very different shapes, such as the flat and square cells of Haloquadratum walsbyi. Despite this morphological similarity to bacteria, archaea possess genes and several metabolic pathways that are more closely related to those of eukaryotes, notably for the enzymes involved in transcription and translation. Other aspects of archaeal biochemistry are unique, such as their reliance on ether lipids in their cell membranes, including archaeols. Archaea use more energy sources than eukaryotes: these range from organic compounds, such as sugars, to ammonia, metal ions or even hydrogen gas. Salt-tolerant archaea (the Haloarchaea) use sunlight as an energy source, and other species of archaea fix carbon, but unlike plants and cyanobacteria, no known species of archaea does both. Archaea reproduce asexually by binary fission, fragmentation, or budding; unlike bacteria, no known species of Archaea form endospores. The first observed archaea were extremophiles, living in extreme environments, such as hot springs and salt lakes with no other organisms. Improved detection tools led to the discovery of archaea in almost every habitat, including soil, oceans, and marshlands. Archaea are particularly numerous in the oceans, and the archaea in plankton may be one of the most abundant groups of organisms on the planet.
Archaea are a major part of Earth’s life. They are part of the microbiota of all organisms. In the human microbiota, they are important in the gut, mouth, and on the skin. Their morphological, metabolic, and geographical diversity permits them to play multiple ecological roles: carbon fixation; nitrogen cycling; organic compound turnover; and maintaining microbial symbiotic and syntrophic communities, for example.
No clear examples of archaeal pathogens or parasites are known. Instead they are often mutualists or commensals, such as the methanogens (methane-producing strains) that inhabit the gastrointestinal tract in humans and ruminants, where their vast numbers aid digestion. Methanogens are also used in biogas production and sewage treatment, and biotechnology exploits enzymes from extremophile archaea that can endure high temperatures and organic solvents.
4.7.1 Origin And evolution
The age of the Earth is about 4.54 billion years. Scientific evidence suggests that life began on Earth at least 3.5 billion years ago. The earliest evidence for life on Earth is graphite found to be biogenic in 3.7-billion-year-old metasedimentary rocks discovered in Western Greenland and microbial mat fossils found in 3.48-billion-year-old sandstone discovered in Western Australia. In 2015, possible remains of biotic matter were found in 4.1-billion-year-old rocks in Western Australia.
Although probable prokaryotic cell fossils date to almost 3.5 billion years ago, most prokaryotes do not have distinctive morphologies, and fossil shapes cannot be used to identify them as archaea. Instead, chemical fossils of unique lipids are more informative because such compounds do not occur in other organisms. Some publications suggest that archaeal or eukaryotic lipid remains are present in shales dating from 2.7 billion years ago; though such data have since been questioned. These lipids have also been detected in even older rocks from west Greenland. The oldest such traces come from the Isua district, which includes Earth’s oldest known sediments, formed 3.8 billion years ago. The archaeal lineage may be the most ancient that exists on Earth.
Woese argued that the Bacteria, Archaea, and Eukaryotes represent separate lines of descent that diverged early on from an ancestral colony of organisms. One possibility is that this occurred before the evolution of cells, when the lack of a typical cell membrane allowed unrestricted lateral gene transfer, and that the common ancestors of the three domains arose by fixation of specific subsets of genes. It is possible that the last common ancestor of bacteria and archaea was a thermophile, which raises the possibility that lower temperatures are “extreme environments” for archaea, and organisms that live in cooler environments appeared only later. Since archaea and bacteria are no more related to each other than they are to eukaryotes, the term prokaryote may suggest a false similarity between them. However, structural and functional similarities between lineages often occur because of shared ancestral traits or evolutionary convergence. These similarities are known as a grade, and prokaryotes are best thought of a grade of life, characterized by such features as an absence of membrane-bound organelles.
The following table (4.2 compares some major characteristics of the three domains, to illustrate their similarities and differences.
| Property | Archaea | Bacteria | Eukarya |
|---|---|---|---|
| Cell membrane | Ether-linked lipids | Ester-linked lipids | Ester-linked lipids |
| Cell wall | Pseudopeptidoglycan, glycoprotein, or S-layer | Peptidoglycan, S-layer, or no cell wall | Various structures |
| Gene structure | Circular chromosomes, similar translation and transcription to Eukarya | Circular chromosomes, unique translation and transcription | Multiple, linear chromosomes, but translation and transcription similar to Archaea |
| Internal cell structure | No membrane-bound organelles (?[63]) or nucleus | No membrane-bound organelles or nucleus | Membrane-bound organelles and nucleus |
| Metabolism[64] | Various, including diazotrophy, with methanogenesis unique to Archaea | Various, including photosynthesis, aerobic and anaerobic respiration, fermentation, diazotrophy, and autotrophy | Photosynthesis, cellular respiration, and fermentation; no diazotrophy |
| Reproduction | Asexual reproduction, horizontal gene transfer | Asexual reproduction, horizontal gene transfer | Sexual and asexual reproduction |
| Protein synthesis initiation | Methionine | Formylmethionine | Methionine |
| RNA polymerase | Many | One | Many |
| EF-2/EF-G | Sensitive to diphtheria toxin | Resistant to diphtheria toxin | Sensitive to diphtheria toxin |
Archaea were split off as a third domain because of the large differences in their ribosomal RNA structure. The particular molecule 16S rRNA is key to the production of proteins in all organisms. Because this function is so central to life, organisms with mutations in their 16S rRNA are unlikely to survive, leading to great (but not absolute) stability in the structure of this nucleotide over generations. 16S rRNA is large enough to show organism-specific variations, but still small enough to be compared quickly. In 1977, Carl Woese, a microbiologist studying the genetic sequences of organisms, developed a new comparison method that involved splitting the RNA into fragments that could be sorted and compared with other fragments from other organisms. The more similar the patterns between species, the more closely they are related.
Woese used his new rRNA comparison method to categorize and contrast different organisms. He compared a variety of species and happened upon a group of methanogens with rRNA vastly different from any known prokaryotes or eukaryotes. These methanogens were much more similar to each other than to other organisms, leading Woese to propose the new domain of Archaea. His experiments showed that the archaea were genetically more similar to eukaryotes than prokaryotes, even though they were more similar to prokaryotes in structure. This led to the conclusion that Archaea and Eukarya shared a common ancestor more recent than Eukarya and Bacteria. The development of the nucleus occurred after the split between Bacteria and this common ancestor.
One property unique to archaea is the abundant use of ether-linked lipids in their cell membranes. Ether linkages are more chemically stable than the ester linkages found in bacteria and eukarya, which may be a contributing factor to the ability of many archaea to survive in extreme environments that place heavy stress on cell membranes, such as extreme heat and salinity. Comparative analysis of archaeal genomes has also identified several molecular conserved signature indels and signature proteins uniquely present in either all archaea or different main groups within archaea. Another unique feature of archaea, found in no other organisms, is methanogenesis (the metabolic production of methane). Methanogenic archaea play a pivotal role in ecosystems with organisms that derive energy from oxidation of methane, many of which are bacteria, as they are often a major source of methane in such environments and can play a role as primary producers. Methanogens also play a critical role in the carbon cycle, breaking down organic carbon into methane, which is also a major greenhouse gas.
4.7.2 Relationship To Bacteria
The relationships among the three domains are of central importance for understanding the origin of life. Most of the metabolic pathways, which are the object of the majority of an organism’s genes, are common between Archaea and Bacteria, while most genes involved in genome expression are common between Archaea and Eukarya. Within prokaryotes, archaeal cell structure is most similar to that of gram-positive bacteria, largely because both have a single lipid bilayer and usually contain a thick sacculus (exoskeleton) of varying chemical composition. In some phylogenetic trees based upon different gene/protein sequences of prokaryotic homologs, the archaeal homologs are more closely related to those of gram-positive bacteria. Archaea and gram-positive bacteria also share conserved indels in a number of important proteins, such as Hsp70 and glutamine synthetase I; but the phylogeny of these genes was interpreted to reveal interdomain gene transfer, and might not reflect the organismal relationship(s).
It has been proposed that the archaea evolved from gram-positive bacteria in response to antibiotic selection pressure. This is suggested by the observation that archaea are resistant to a wide variety of antibiotics that are produced primarily by gram-positive bacteria, and that these antibiotics act primarily on the genes that distinguish archaea from bacteria. The proposal is that the selective pressure towards resistance generated by the gram-positive antibiotics was eventually sufficient to cause extensive changes in many of the antibiotics’ target genes, and that these strains represented the common ancestors of present-day Archaea. The evolution of Archaea in response to antibiotic selection, or any other competitive selective pressure, could also explain their adaptation to extreme environments (such as high temperature or acidity) as the result of a search for unoccupied niches to escape from antibiotic-producing organisms; Cavalier-Smith has made a similar suggestion. This proposal is also supported by other work investigating protein structural relationships and studies that suggest that gram-positive bacteria may constitute the earliest branching lineages within the prokaryotes.
4.7.3 Relation To Eukaryotes
The evolutionary relationship between archaea and eukaryotes remains unclear. Aside from the similarities in cell structure and function that are discussed below, many genetic trees group the two.
Complicating factors include claims that the relationship between eukaryotes and the archaeal phylum Crenarchaeota is closer than the relationship between the Euryarchaeota and the phylum Crenarchaeota and the presence of archaea-like genes in certain bacteria, such as Thermotoga maritima, from horizontal gene transfer. The standard hypothesis states that the ancestor of the eukaryotes diverged early from the Archaea, and that eukaryotes arose through fusion of an archaean and eubacterium, which became the nucleus and cytoplasm; this hypothesis explains various genetic similarities but runs into difficulties explaining cell structure. An alternative hypothesis, the eocyte hypothesis, posits that Eukaryota emerged relatively late from the Archaea.
A lineage of archaea discovered in 2015, Lokiarchaeum (of proposed new Phylum “Lokiarchaeota”), named for a hydrothermal vent called Loki’s Castle in the Arctic Ocean, was found to be the most closely related to eukaryotes known at that time. It has been called a transitional organism between prokaryotes and eukaryotes.
Several sister phyla of “Lokiarchaeota” have since been found (“Thorarchaeota”, “Odinarchaeota”, “Heimdallarchaeota”), all together comprising a newly proposed supergroup Asgard, which may appear as a sister taxon to Proteoarchaeota.
Details of the relation of Asgard members and eukaryotes are still under consideration, although, in January 2020, scientists reported that Candidatus Prometheoarchaeum syntrophicum, a type of Asgard archaea, may be a possible link between simple prokaryotic and complex eukaryotic microorganisms about two billion years ago.
4.7.4 Morphology
Individual archaea range from 0.1 micrometers (μm) to over 15 μm in diameter, and occur in various shapes, commonly as spheres, rods, spirals or plates. Other morphologies in the Crenarchaeota include irregularly shaped lobed cells in Sulfolobus, needle-like filaments that are less than half a micrometer in diameter in Thermofilum, and almost perfectly rectangular rods in Thermoproteus and Pyrobaculum. Archaea in the genus Haloquadratum such as Haloquadratum walsbyi are flat, square specimens that live in hypersaline pools. These unusual shapes are probably maintained by both their cell walls and a prokaryotic cytoskeleton. Proteins related to the cytoskeleton components of other organisms exist in archaea, and filaments form within their cells, but in contrast with other organisms, these cellular structures are poorly understood. In Thermoplasma and Ferroplasma the lack of a cell wall means that the cells have irregular shapes, and can resemble amoebae.
Some species form aggregates or filaments of cells up to 200 μm long. These organisms can be prominent in biofilms. Notably, aggregates of Thermococcus coalescens cells fuse together in culture, forming single giant cells. Archaea in the genus Pyrodictium produce an elaborate multicell colony involving arrays of long, thin hollow tubes called cannulae that stick out from the cells’ surfaces and connect them into a dense bush-like agglomeration. The function of these cannulae is not settled, but they may allow communication or nutrient exchange with neighbors. Multi-species colonies exist, such as the “string-of-pearls” community that was discovered in 2001 in a German swamp. Round whitish colonies of a novel Euryarchaeota species are spaced along thin filaments that can range up to 15 centimetres (5.9 in) long; these filaments are made of a particular bacteria species.
4.7.5 Structure, Composition Development, And Operation
Archaea and bacteria have generally similar cell structure, but cell composition and organization set the archaea apart. Like bacteria, archaea lack interior membranes and organelles. Like bacteria, the cell membranes of archaea are usually bounded by a cell wall and they swim using one or more flagella. Structurally, archaea are most similar to gram-positive bacteria. Most have a single plasma membrane and cell wall, and lack a periplasmic space; the exception to this general rule is Ignicoccus, which possess a particularly large periplasm that contains membrane-bound vesicles and is enclosed by an outer membrane.
4.7.6 Cell Wall And Flagella
Most archaea (but not Thermoplasma and Ferroplasma) possess a cell wall. In most archaea the wall is assembled from surface-layer proteins, which form an S-layer. An S-layer is a rigid array of protein molecules that cover the outside of the cell (like chain mail). This layer provides both chemical and physical protection, and can prevent macromolecules from contacting the cell membrane. Unlike bacteria, archaea lack peptidoglycan in their cell walls. Methanobacteriales do have cell walls containing pseudopeptidoglycan, which resembles eubacterial peptidoglycan in morphology, function, and physical structure, but pseudopeptidoglycan is distinct in chemical structure; it lacks D-amino acids and N-acetylmuramic acid, substituting the latter with N-Acetyltalosaminuronic acid.
Archaeal flagella are known as archaella, that operate like bacterial flagella – their long stalks are driven by rotatory motors at the base. These motors are powered by a proton gradient across the membrane, but archaella are notably different in composition and development. The two types of flagella evolved from different ancestors. The bacterial flagellum shares a common ancestor with the type III secretion system, while archaeal flagella appear to have evolved from bacterial type IV pili. In contrast with the bacterial flagellum, which is hollow and assembled by subunits moving up the central pore to the tip of the flagella, archaeal flagella are synthesized by adding subunits at the base.
4.7.7 Membranes
Archaeal membranes are made of molecules that are distinctly different from those in all other life forms, showing that archaea are related only distantly to bacteria and eukaryotes. In all organisms, cell membranes are made of molecules known as phospholipids. These molecules possess both a polar part that dissolves in water (the phosphate “head”), and a “greasy” non-polar part that does not (the lipid tail). These dissimilar parts are connected by a glycerol moiety. In water, phospholipids cluster, with the heads facing the water and the tails facing away from it. The major structure in cell membranes is a double layer of these phospholipids, which is called a lipid bilayer.
The phospholipids of archaea are unusual in four ways:
- They have membranes composed of glycerol-ether lipids, whereas bacteria and eukaryotes have membranes composed mainly of glycerol-ester lipids. The difference is the type of bond that joins the lipids to the glycerol moiety; the two types are shown in yellow in the figure at the right. In ester lipids this is an ester bond, whereas in ether lipids this is an ether bond.
- The stereochemistry of the archaeal glycerol moiety is the mirror image of that found in other organisms. The glycerol moiety can occur in two forms that are mirror images of one another, called enantiomers. Just as a right hand does not fit easily into a left-handed glove, enantiomers of one type generally cannot be used or made by enzymes adapted for the other. The archaeal phospholipids are built on a backbone of sn-glycerol-1-phosphate, which is an enantiomer of sn-glycerol-3-phosphate, the phospholipid backbone found in bacteria and eukaryotes. This suggests that archaea use entirely different enzymes for synthesizing phospholipids as compared to bacteria and eukaryotes. Such enzymes developed very early in life’s history, indicating an early split from the other two domains.
- Archaeal lipid tails differ from those of other organisms in that they are based upon long isoprenoid chains with multiple side-branches, sometimes with cyclopropane or cyclohexane rings. By contrast, the fatty acids in the membranes of other organisms have straight chains without side branches or rings. Although isoprenoids play an important role in the biochemistry of many organisms, only the archaea use them to make phospholipids. These branched chains may help prevent archaeal membranes from leaking at high temperatures.
- In some archaea, the lipid bilayer is replaced by a monolayer. In effect, the archaea fuse the tails of two phospholipid molecules into a single molecule with two polar heads (a bolaamphiphile); this fusion may make their membranes more rigid and better able to resist harsh environments. For example, the lipids in Ferroplasma are of this type, which is thought to aid this organism’s survival in its highly acidic habitat.
4.7.8 Archaeal Metabolism
Archaea exhibit a great variety of chemical reactions in their metabolism and use many sources of energy. These reactions are classified into nutritional groups, depending on energy and carbon sources. Some archaea obtain energy from inorganic compounds such as sulfur or ammonia (they are chemotrophs). These include nitrifiers, methanogens and anaerobic methane oxidisers. In these reactions one compound passes electrons to another (in a redox reaction), releasing energy to fuel the cell’s activities. One compound acts as an electron donor and one as an electron acceptor. The energy released is used to generate adenosine triphosphate (ATP) through chemiosmosis, the same basic process that happens in the mitochondrion of eukaryotic cells.
Other groups of archaea use sunlight as a source of energy (they are phototrophs), but oxygen–generating photosynthesis does not occur in any of these organisms. Many basic metabolic pathways are shared among all forms of life; for example, archaea use a modified form of glycolysis (the Entner–Doudoroff pathway) and either a complete or partial citric acid cycle. These similarities to other organisms probably reflect both early origins in the history of life and their high level of efficiency.
| Nutritional Type | Source of Energy | Source of Carbon | Examples |
|---|---|---|---|
| Phototrophs | Sunlight | Organic compounds | Halobacterium |
| Lithotrophs | Inorganic compounds | Organic compounds or carbon fixation | Ferroglobus, Methanobacteria or Pyrolobus |
| Organotrophs | Organic compounds | Organic compounds or carbon fixation | Pyrococcus, Sulfolobus or Methanosarcinales |
Some Euryarchaeota are methanogens (archaea that produce methane as a result of metabolism) living in anaerobic environments, such as swamps. This form of metabolism evolved early, and it is even possible that the first free-living organism was a methanogen. A common reaction involves the use of carbon dioxide as an electron acceptor to oxidize hydrogen. Methanogenesis involves a range of coenzymes that are unique to these archaea, such as coenzyme M and methanofuran. Other organic compounds such as alcohols, acetic acid or formic acid are used as alternative electron acceptors by methanogens. These reactions are common in gut-dwelling archaea. Acetic acid is also broken down into methane and carbon dioxide directly, by acetotrophic archaea. These acetotrophs are archaea in the order Methanosarcinales, and are a major part of the communities of microorganisms that produce biogas.
Other archaea use CO2 in the atmosphere as a source of carbon, in a process called carbon fixation (they are autotrophs). This process involves either a highly modified form of the Calvin cycle or another metabolic pathway called the 3-hydroxypropionate/ 4-hydroxybutyrate cycle. The Crenarchaeota also use the reverse Krebs cycle while the Euryarchaeota also use the reductive acetyl-CoA pathway. Carbon fixation is powered by inorganic energy sources. No known archaea carry out photosynthesis. Archaeal energy sources are extremely diverse, and range from the oxidation of ammonia by the Nitrosopumilales to the oxidation of hydrogen sulfide or elemental sulfur by species of Sulfolobus, using either oxygen or metal ions as electron acceptors.
Phototrophic archaea use light to produce chemical energy in the form of ATP. In the Halobacteria, light-activated ion pumps like bacteriorhodopsin and halorhodopsin generate ion gradients by pumping ions out of and into the cell across the plasma membrane. The energy stored in these electrochemical gradients is then converted into ATP by ATP synthase. This process is a form of photophosphorylation. The ability of these light-driven pumps to move ions across membranes depends on light-driven changes in the structure of a retinol cofactor buried in the center of the protein.
4.7.9 Archaeal Genetics
Archaea usually have a single circular chromosome, with as many as 5,751,492 base pairs in Methanosarcina acetivorans, the largest known archaeal genome. The tiny 490,885 base-pair genome of Nanoarchaeum equitans is one-tenth of this size and the smallest archaeal genome known; it is estimated to contain only 537 protein-encoding genes. Smaller independent pieces of DNA, called plasmids, are also found in archaea. Plasmids may be transferred between cells by physical contact, in a process that may be similar to bacterial conjugation.
Archaea are genetically distinct from bacteria and eukaryotes, with up to 15% of the proteins encoded by any one archaeal genome being unique to the domain, although most of these unique genes have no known function. Of the remainder of the unique proteins that have an identified function, most belong to the Euryarchaea and are involved in methanogenesis. The proteins that archaea, bacteria and eukaryotes share form a common core of cell function, relating mostly to transcription, translation, and nucleotide metabolism. Other characteristic archaeal features are the organization of genes of related function – such as enzymes that catalyze steps in the same metabolic pathway into novel operons, and large differences in tRNA genes and their aminoacyl tRNA synthetases.
Transcription in archaea more closely resembles eukaryotic than bacterial transcription, with the archaeal RNA polymerase being very close to its equivalent in eukaryotes, while archaeal translation shows signs of both bacterial and eukaryotic equivalents. Although archaea have only one type of RNA polymerase, its structure and function in transcription seems to be close to that of the eukaryotic RNA polymerase II, with similar protein assemblies (the general transcription factors) directing the binding of the RNA polymerase to a gene’s promoter, but other archaeal transcription factors are closer to those found in bacteria. Post-transcriptional modification is simpler than in eukaryotes, since most archaeal genes lack introns, although there are many introns in their transfer RNA and ribosomal RNA genes, and introns may occur in a few protein-encoding genes.
4.7.10 Archaeal Ecology
Archaea exist in a broad range of habitats, and are now recognized as a major part of global ecosystems, and may represent about 20% of microbial cells in the oceans. However, the first-discovered archaeans were extremophiles. Indeed, some archaea survive high temperatures, often above 100 °C (212 °F), as found in geysers, black smokers, and oil wells. Other common habitats include very cold habitats and highly saline, acidic, or alkaline water, but archaea include mesophiles that grow in mild conditions, in swamps and marshland, sewage, the oceans, the intestinal tract of animals, and soils.
Extremophile archaea are members of four main physiological groups. These are the halophiles, thermophiles, alkaliphiles, and acidophiles. These groups are not comprehensive or phylum-specific, nor are they mutually exclusive, since some archaea belong to several groups. Nonetheless, they are a useful starting point for classification.
Halophiles, including the genus Halobacterium, live in extremely saline environments such as salt lakes and outnumber their bacterial counterparts at salinities greater than 20–25%. Thermophiles grow best at temperatures above 45 °C (113 °F), in places such as hot springs; hyperthermophilic archaea grow optimally at temperatures greater than 80 °C (176 °F). The archaeal Methanopyrus kandleri Strain 116 can even reproduce at 122 °C (252 °F), the highest recorded temperature of any organism.
Other archaea exist in very acidic or alkaline conditions. For example, one of the most extreme archaean acidophiles is Picrophilus torridus, which grows at pH 0, which is equivalent to thriving in 1.2 molar sulfuric acid.
This resistance to extreme environments has made archaea the focus of speculation about the possible properties of extraterrestrial life. Some extremophile habitats are not dissimilar to those on Mars, leading to the suggestion that viable microbes could be transferred between planets in meteorites.
Recently, several studies have shown that archaea exist not only in mesophilic and thermophilic environments but are also present, sometimes in high numbers, at low temperatures as well. For example, archaea are common in cold oceanic environments such as polar seas. Even more significant are the large numbers of archaea found throughout the world’s oceans in non-extreme habitats among the plankton community (as part of the picoplankton). Although these archaea can be present in extremely high numbers (up to 40% of the microbial biomass), almost none of these species have been isolated and studied in pure culture. Consequently, our understanding of the role of archaea in ocean ecology is rudimentary, so their full influence on global biogeochemical cycles remains largely unexplored. Some marine Crenarchaeota are capable of nitrification, suggesting these organisms may affect the oceanic nitrogen cycle, although these oceanic Crenarchaeota may also use other sources of energy.
Vast numbers of archaea are also found in the sediments that cover the sea floor, with these organisms making up the majority of living cells at depths over 1 meter below the ocean bottom. It has been demonstrated that in all oceanic surface sediments (from 1000- to 10,000-m water depth), the impact of viral infection is higher on archaea than on bacteria and virus-induced lysis of archaea accounts for up to one-third of the total microbial biomass killed, resulting in the release of ~0.3 to 0.5 gigatons of carbon per year globally.
4.7.11 Significance In Technology And Industry
Extremophile archaea, particularly those resistant either to heat or to extremes of acidity and alkalinity, are a source of enzymes that function under these harsh conditions. These enzymes have found many uses. For example, thermostable DNA polymerases, such as the Pfu DNA polymerase from Pyrococcus furiosus, revolutionized molecular biology by allowing the polymerase chain reaction to be used in research as a simple and rapid technique for cloning DNA. In industry, amylases, galactosidases and pullulanases in other species of Pyrococcus that function at over 100 °C (212 °F) allow food processing at high temperatures, such as the production of low lactose milk and whey. Enzymes from these thermophilic archaea also tend to be very stable in organic solvents, allowing their use in environmentally friendly processes in green chemistry that synthesize organic compounds. This stability makes them easier to use in structural biology. Consequently, the counterparts of bacterial or eukaryotic enzymes from extremophile archaea are often used in structural studies.
In contrast with the range of applications of archaean enzymes, the use of the organisms themselves in biotechnology is less developed. Methanogenic archaea are a vital part of sewage treatment, since they are part of the community of microorganisms that carry out anaerobic digestion and produce biogas. In mineral processing, acidophilic archaea display promise for the extraction of metals from ores, including gold, cobalt and copper.
Archaea host a new class of potentially useful antibiotics. A few of these archaeocins have been characterized, but hundreds more are believed to exist, especially within Haloarchaea and Sulfolobus. These compounds differ in structure from bacterial antibiotics, so they may have novel modes of action. In addition, they may allow the creation of new selectable markers for use in archaeal molecular biology.